POSTER SESSION
Alfalfa Diversity in Mexico - L.
Castro-Acero,
J. J. Marquez-Ortiz, and J. Santamaria-Cesar..22
Alfalfa On-Farm Research at North-Central
Mexico - J. J. Marquez O., G. Nunez H.,
Y. I. Chew M., E. Castro M. and M. Ramirez
D .......................................23
The Yield and Quality of the U. S.
A. Multileaf Alfalfa Varieties in Poland
G. Harasimowicz-Hermann, 1. Andrzejewska,
W. Nowak,
F. Gospodarcyk and W. Waniorek .......................................................24
The Improvement of Alfalfa Breeding in China
- Zhensheng Gao, Xueju Xie,
Qidong Ma and Fuzeng Hong...............................................................25
The Study on Isozymic of Local Alfalfa Cultivars
in China - Xueju Xie,
Mojun Chen and Zhensheng Gao............................................................26
Genetic Diversity in Wild Accessions of Medicago
sativa from Spain
E. Jenczewski, G. Genier, J. Ronfort and J. M.
Prosperi ................................27
Effect of Grazing Frequency and Intensity
on Yield and Persistence of Alfalfa
Tall Fescue Pastures - Nestor A. Romero and
N. Tony Juan ...............................28
Inbreeding as a Method of Developing Homoygous
Lines in Alfalfa, Medicago
sativa- E. V. Kvasova ........................................................................29
WEEDALF$: An Integrated Expert System for Weed
Management in Alfalfa
J. F. Stritzke, C. W. Cuperus, and S. Zhang............................................30
Breeding a Bloat-Tolerant Alfalfa in Argentina
- D. H. Basigalup,
C. V. Castell and C. D. Giaveno .......................................................31
Genetic Variation for Digestibility and Fiber Contents
in the Medicago sativa
Complex - B. Julier and C. Huyghe ...............................................32
Biotechnological Improvement of Alfalfa Nutritive
Quality by Anti-sense Expression
of COMT and CCOMT, Methylating Enzymes in Lignin
Biosynthesis
V. J. H. Sewalt, J. W. Blount, and R. A. Dixon
...................................33
Alfalfa Yield Response to Method and Rate of
Applied P and Weed Intensity
S. B. Phillips, G. V. Johnson, and W. R. Raun
....................................34
Aluminum Tolerance QTL in Diploid Alfalfa - M.
K. Sledge, J. H. Bouton,
J. Tamulonis, G. Kochert, and W. A. Parrot .......................................35
Predicting Acid/Aluminum Tolerance in Alfalfa
Genotypes with Hematoxylin
Staining - Kelley, Rowena Y., Powell, D.;
Yang, G.; Glass, M......................36
Cis-elements and Trans-acting Factors Required for
the Regulation of Alfalfa
Isoflavone Reductase - B. Miao and N. Paiva.......................................37
Toward Metabolic Engineering of Alfalfa for Production
of Prenylated Isoflavonoid/
Pterocarpan Phytoalexins - Z. Guo and N. L. Paiva.................................38
Alfagenes - A Comprehensive Internet-accessible
ACE Database for Alfalfa; http://
probe.nalusda.gov - D. Z. Skinner, P. C. St. Amand,
and S. M. Ramaiah.............39
Risk of Alfalfa Transgene Dissemination and
Scale Dependent Effects - P. C. St.
Amand, D. Z. Skinner, and R. N. Peaden ...........................................40
Mapping of Simple Sequence Repeats (SSR) DNA Markers
in Diploid and Tetraploid
Alfalfa - Noa Diwan, Joseph H. Bouton, Gary Kochert,
Arvind A.
Bhagwat, and Perry B. Cregan .....................................................41
Divergent Selection for Self-fertility in Alfalfa
and Correlated Responses for Forage
Yield - D. Rosellini, F. Veronesi and G. Barcaccia................................42
Ploidy Determination of Medicago sativa ssp.
falcata germplasm: Step I in a falcata
germplasm enhancement program - E. C. Brummer
and S. Sin..........................43
Progeny Test Based on Morphological and Molecular
Markers to Verify the
Occurrence of Parthenogenesis in an Apomeiotic
Mutant of Diploid
Alfalfa - G. Barcaccia, S. Tavoletti, M. Falcinelli
and F. Veronesi ..............44
Assessment of Genetic Variability for Cryoprotective
Oligosaccharide Accumulation
in Cold-acclimated Leaves of Alfalfa - Y. Castonguay,
P. Nadeau and R. Michaud...45
Analysis of Seed Storage Proteins in Regenerated
Plants of Alfalfa (Medicago varia Mart.)
O. Tzeveleva, S. Peltek, E. Deineko .............................................46
Callus Initiation and Development of Medicago
sativa L. Cultured in vitro
Guochen Yang and Marihelen Kamp-Glass .....................................47
Molecular Genetics of a Model-Plant: Medicago truncatula
- T. Huguet, M. Gherardi,
L. Tirichine, I. Bonnin, G. Genier, J. Ronfort, J.
M. Prosperi...................48
Evaluation of Cold Tolerance in Annual Medics
with Potential for Use in Rotation
with wheat on the U. S. High Plains - J. M. Krall,
R. H. Delaney
D. A. Claypool and R. W. Groose.....................................................49
Breeding Medicago lupulina for Pasture
and Green Manure - G. V. Stepanova,
E. N. Atlakova, G. P. Zatchina......................................................50
Assignment of Medicago rigidula Accessions
in the NPGS into the Two Species
M. rigidula and M. rigiduloides - D.
C. Heft and R. W. Groose .............51
Sources of Resistance to Anthracnose in the
Annual Medicago Core Collection
N. R. O'Neill and G. R. Bauchan....................................................52
Influence of Light on Resistance to Vascular
Wilt Diseases of Alfalfa
B. W. Pennypacker, M. L. Risius, J. J. Volenec,
and S. M. Cunningham................53
Persistence and Levels of Crown and Root Rot Diseases
in Alfalfa Cultivars Under Mowing
and Rotational Grazing Management Systems - A. Navarro
and E. H. Hijano ............54
Post-harvest Fungal Resistance in Alfalfa: Cultivar
Response and Mechanisms
V. I. Babij, K. M. Wittenberg, and S. R. Smith,
Jr......................................55
Resistance in the Annual Medicago Core
Collection to Two Isolates of the Downy
Mildew Fungus from Alfalfa - J. R. Yaege and D.
L. Stuteville.......................56
Molecular Markers Associated with Resistance
to Downy Mildew in Tetraploid
Alfalfa - D. E. Obert, D. Z. Skinner, and D. L.
Stuteville...............................57
Dynamics of Apothecial Populations of Sclerotinia
trifoliorum - P. Vincelli,
J. C. Doney, Jr., and L. Wang.............................................................58
Disease Resistance of Australian Lucerne Cultivars
- G. C. Auricht..........................59
Alfalfa Germplasm Resources and Evaluation of
their Disease Resistance in Semi
arid Areas of Gansu Province, People's Republic of
China - Ma Zhengyu...............60
Detection of Antixenosis Resistance Mechanism
to Blue Aphid in an Argentine
Alfalfa Population -J. O. Gieco, D. H. Basigalup
and E. H. Hijano ..................61
Are Chemical Residues Sufficient in Killing
or Deterring Blister Beetle Swarms
in Alfalfa? - P. G. Mulder, Jr. And K. T. Shelton
..................................62
Alfalfa Diversity in Mexico
L. Castro-Acero1, J.J. Marquez-Ortiz2, and J. Santamaria-Cesar2
National Inst. of Forestry, Agriculture and Livestock Research (INIFAP)
1 C.E. Valle de Mexico, Apdo. Postal 10, CP 56230 Chapingo, Mex., Mexico.
2 C.E La Laguna, Apdo. Postal 247, CP 27000 Torreon, Coah., Mexico.
Alfalfa (Medicago sativa L.) was introduced in Mexico from Spain in
the 16th century. Four hundred years of natural selection resulted in Mexican
alfalfa ecotypes. Cultivars selected from Mexican ecotypes have good local
adaptation and higher winter yields than cultivars from California or Spain
(Castro-Acero, 1978). In the 1970s the National Institute for Forestry,
Agriculture and Livestock Research (INIFAP) conducted trials of Mexican
germplasm collections to asses their productivity. Data from these trials
are a valuable basis for designing further studies involving collection
and characterization of alfalfa germplasm in Mexico. The objective of this
research was to characterize and classify Mexican alfalfa germplasm based
on agronomic traits.
The experiment was established in January 1976 at the Valle de Mexico Experiment
Station in Chapingo, Mex. The trial included 46 ecotypes from Atlixco,
Pue.; Atoyac, Jal.; Navojoa, Son.; Oaxaca, Oax.; San Miguel Octopan, Gto.;
Tanhuato, Mich; and Zacapu, Mich. Control cultivars were Moapa and San
Joaquin II (California) and Aragon (Spain). Accesions were evaluated under
a 7 x 7 lattice design with four replications. The experimental unit consisted
of three 5-m rows separated at 30 cm. Twenty four harvests were taken during
a three-year period. We measured dry matter yield; symptoms of leaf spot,
downy mildew, virus, spotted aphid, and pea aphid; plant height; uniformity;
leafiness; regrowth height; plant vigor; and plant survival. Variance and
correlation analyses were used to select classification variables. Variable
standarization, principal component analysis, and average linkage cluster
analysis were used to determine groups based on dendrogram interpretation.
Ecotypes had significant effects (P<0.05) for all variables. We found
six different clusters (number of ecotypes in parenthesis): Zacapu+Atlixco
(15), Zacapu+San Miguel (6), Oaxaca-1 (11), Tanhuato (13), Oaxaca-2 (1),
and Navojoa (3). Clusters Oaxaca-1 and -2, Tanhuato and Navojoa coincided
well with their area of collection. The cluster Tanhuato had high yield,
low virus and aphid damage, and highest plant height, regrowth, vigor and
plant survival scores. Moapa and San Joaquin II fell within Oaxaca-1, a
cluster with low adaptation, suggesting that California cultivars were
as low in adaptation as ecotypes from Oaxaca. Cultivar Aragon was similar
to ecotypes from an array of locations in central Mexico, indicating that
such ecotypes are still similar to those from Spain. This research suggests
that local ecotypes have potential to enhance agronomic performance of
alfalfa in Mexico.
References
Castro-Acero, L. 1978. Alfalfa. p. 165 In T. Cervantes S. (ed.) Recursos
geneticos disponibles a Mexico. SOMEFI. Chapingo, Mex.
Alfalfa On-Farm Research at North-Central
Mexico
J.J. Marquez O., G. Nunez H., Y.I. Chew M., E. Castro M. and M. Ramirez
D.
National Inst. of Forestry, Agriculture and Livestock Research (INIFAP)
C.E La Laguna, Apdo. Postal 247, CP 27000 Torreon, Coah., Mexico.
Alfalfa is the main forage for dairy farms in the temperate and arid
zones of Mexico, where 290 000 ha are grown under irrigation. La Laguna
has the most advanced dairy industry in North-Central Mexico, but, due
to water availability problems, the amount of alfalfa grown in this region
has never surpassed 30 000 ha. Recently, local farmers created La Laguna
Foundation for Agricultural and Livestock Research (PIAL). PIAL coordinates
and supports private (LALA, Inc.) and public (INIFAP) research efforts
to increase forage crop productivity. Integrated Alfalfa Management, a
current project supported by PIAL, involves an on-farm component to promote
and increase technology adoption and design research according to farmer
s needs. Our objective is to develop an integrated management system to
increase alfalfa productivity and persistence under our local environmental
conditions.
In 1995, our research involved introduction of alfalfa nondormant cultivars;
diagnose and control of diseases, insects, and weeds; and postharvest management.
Ten cultivars were evaluated (1 x 3 m plots) in replicated field trials
at three locations. Twenty farms were monitored for disease incidence in
March, June, August, and November. Ten farms were monitored for beneficial
and damaging insects. Six preemergent and seven postemergent herbicide
treatments were tested in two separate trials at one location to determine
their ability to control dodder ( Cuscuta sp). Alfalfa, as hay or silage,
with or without monensin, was fed to cows to assess treatment effect on
dry matter intake during the hot summer season.
Cultivars had a significant (P<0.05) effect on first year fresh and
dry matter yield of alfalfa. The highest yielding cultivar was SW-14; the
lowest, Nitro and Aragon. Diseases were crown rot (Fusarium, Rhizoctonia,
Phoma, Colletotrichum), anthracnose (Colletotrichum), and root rot (Phymatotrichum
omnivorum). Crown rot was significantly (P<0.05) correlated with stand
age. Nematodes (Pratylenchus, Helicotylenchus, Tylenchorynchus, Xiphinema)
were not a serious problem. Damaging insects found included aphids (Therioaphis,
Acyrthosiphon), leafhoppers (Empoasca), Diabrotica, thrips (Frankliniella)
and false bug (Lygus). Beneficial insects were minute pirate bug (Orius),
ladybug (Hippodamia), bigeyed bug Geocoris and Braconidae. Preemergent
herbicides TR-10 (10-20 kg/ha) and Prowl-400 (3-6 l/ha) controlled dodder
efficiently; postemergent herbicides did not. Forage losses were smaller
for silage (13.4%) than for hay (33.9%). Dry matter intake and milk production
and quality did not differ when cows were fed either silage or hay with
or without monensin.
New research involves agronomic and nutritive characterization of germplasm
available in Mexico, estimation of alfalfa losses caused by crown rot,
and screening germplasm for local diseases.
The Yield and Ouality of the U.S.A.
Multileaf Alfalfa Varieties In Poland
G. Harasimowicz-Hermann*, J. Andrzejewska*, W. Nowak**, F. Gospodarczyk**
and W. Waniorek***
*University of Technology and Agriculture, Mazowiecka St., 8S-084 Bydgoszcz
**University of Agriculture, Norwid St., 50-375 Wroclaw
***Agricultural Experiment Station, 64-312 Brody, Poland
The objective of the study was to determine the usefulness of U.S.A.
multileaf alfalfa varieties for cultivation under varied Polish soil and
climatic conditions. Multileaf forms differ from the others because up
to 30% their leaves consist of 4-7 leaflets instead of a typical trifolium.
Alfalfa variety 'Legend' and the genera FG3B50 and FG4B59, breeded by Dr.
Mark McCaslin, Forage Genetics, West Salem, WI, were tested. Two series
of three-year-long multipoint field experiments were settled in 1993 and
1994. The U.S.A. varieties were compared with the standard Polish variety
'Radius', well prepared for Polish conditions. Alfalfa yields of green
and dry matter were assayed and evaluated statistically. Total protein,
fiber and soluble sugars content, digestibility in vitro, winterhardiness,
regrowth rate, as well as stand density were also assessed. Determinations
made for I series (1993-95) include the seeding ycar, and two production
years, while those made for II series (1994-95) the seeding year and first
production year. The highest yields of green and dry matter and protein
were found for I series at Brody where alfalfa varieties were seeded alone.
The experiment was conducted on brown soil developed from boulder clay.
Average rainfall for that period was 765 mm and the temperature 8.5 deg
C. High yields were also noted near Wroclaw, where alfalfa was cultivated
on brown soil of typical grey-brown podzolic type. In the period of study
average rainfall was 1750mm and the temperature 8.5 deg C. Most unfavourable
soil and climatic conditions occur at the experiment site ncar Bydgoszcz,
where the lowest alfalfa yields were observed. The soil type is pseudo
podzolic on strong loamy sand. The average rainfall was 443 mm and the
temperature 8.3 deg C. The results of I series indicated that the level
of yields and forage quality of multileaf 'Legend' were close to those
of the standard 'Radius', while 'Legend' was significantly better in II
series. In the seeding year higher yields, especially in case of green
matter, were noted for 'Legend'. Very good soil, moisture and thermal conditions
at Brody promoted high and equalized yields of all the varieties studied.
In production years the climate of south-west Poland caused better performance
of the U.S.A. varieties, especially 'Legend'. Under most difficult soil
and climatic conditions of central Poland 'Radius' yielded higher in second
production year than the U.S.A. varieties. Total protein yields (I series)
for 'Radius' and 'Legend' were 4345 and 4370 kg/ha, respectively, while
the corresponding sequence for II series was: 'Legend'>FG3B50>Radius>FG4B59
(2253, 2043, 1997, 1994 kg/ha, respectively). Fodder quality of all varieties
was similar, with one exception for FG4B59, for which- the highest protein
yield was found. Changing soil and climatic conditions did not affect the
fodder quality. Winterhardiness and regrowth rate ofthe U.S.A. varieties
did not vary from the Polish one.
The Improvement of alfalfa Breeding in China
Zhensheng Gao, Xueju Xie, Qidong Ma and Fuzeng Hong
Grassland Institute, China Agricultural University, Beijing 100094, China
Alfalfa is the most widespread cultivated and applied legumes around
the world which had the highest utilization value. China has more than
2,000 years history in introduction of alfalfa, but breeding and selection
of new varieties is a relatively new event in China. The selection and
introduction of important herbage cultivars was started in the fifties
in order to advocate the rotation of grassland crops and to develop the
animal husbandry, such as 'Gongnong No.l' and 'Gongnong No.2' Medicago
sativa L., which was bred by single and mixed selection method, based on
the introduction of 'Grimm' alfalfa at the end of fifties. In the seventies,
researchers used local M. sativa L. cross wild M. falcata to enhance the
cold tolerance and drought resistance, bred 'Caoyuan No. I' and 'Caoyuan
No.2' M. varia Martin. with the started working on Chinese herbage cultivar
licensing and registration since 1986, researchers has compared and evaluated
the alfalfa around China, which had appropriated to grow in the local areas
after long time of adaptation, collection and collation a series of local
alfalfa varieties.
There are twelve species and three formas in the genus, which are grown
in China. The most commonly grown species are Medicago sativa L., M. falcata
L., M. media Pers, M. ruthenica L., M. hispida Gaerth. and M. lupulina
L. M. sativa and M. hispida is common]y grown in the south of China. Nowadays,
China has totally 30 licensed alfalfa cultivars, including 23 M. sativa
L. cultivars and 7 M. varia L. cultivars.
Chinese Herbage Cultivar Registration Board (CHCRB) was established
in 1987. From 1986 to 1994, CHCRB has registered totally 163 herbage, fodder
crops and turfgrasses, including 78 grasses, 70 legumes and 15 other families.
The proportion of alfalfa cultivars in total registered cultivars is 18.4%
and 38.5% in total legumes cultivars, including 11 newly bred cultivars,
17 local cultivars, 1 wild species and I introduced cultivar. Most ofthe
17 local cultivars are M. sativa L. There are 1.3 million hectares alfalfa
plant areas in Chilla, mainly distributed in Northwest 66.8%, North China
30.3% and Northeast 2.7%, licensed cultivars in Northwest is 40%, 30% in
North China and 23% in Northeast of China.
References
1. Edited by the Chinese Herbage Cultivar Registration Board (CHCRB), 1992,
LICENSED CULTIVARS OF HERBAGE CROPS IN CHINA, Beijing Agricultural University
Press, Beijing, China.
2. Edited by CHCRB, 1995, Working report of Chinese Herbage cultivar Registration
Board.
The Study on Isozymic of Local Alfalfa Cultivars
in China
Xueju Xie, Mojun Chell and Zhenshellg Gao
Grassland Institute, China Agricultural University, Beijing 100094, China
Alfalfa were introduced into China about two thousands years ago. It
was widely spread through out provinces and regions of north of the Changjiang
River. The sources of local alfalfa cultivars are very rich in China. Thirty
alfalfa cultivars were licensed by the Chinese Herbage Registration Board
at the end of 1995, including 17 local cultivars which belong to the M.
sativa L.. In the palmer, we collected total 17 local cultivars and domesticated
wild cultivar (M. media Pers.). Esterase isozyme of single plant of every
cultivar was analyzed, in order to find out the esterase isozyme band -l
types, and the relationship between different cultivars. th result showed:
total 14 isozyme bands can be obtained in four leaves stage. Among them,
7A, 12A, 13A, and 14A are the public bands of alfalfa. But the emerge frequency
of other bands is different in every cultivar. Correlation analysis on
11 bands and 7 meteorological factors showed that the frequency of 2A,
3A, 4A, 5A and 8A had correlation with average year temperature, average
lowest temperature, average highest temperature and >5 °C year temperature
summation.
Statistical analysis of esterase isozyme of 900 materials from single
plant of every cultivar showed that Est-zymogram types have 60 patterns,
32 patterns are specific types of cultivars. Est-zymogram types are not
only varied, but also abundant among cultivars and different plants of
every cultivar. Experimental evidences indicate that the gene resources
of alfalfa are very rich in China.
Cluster analysis on esterase isozyme of 18 cultivars showed that relationship
was close among Pianguan, Hexi, Aletai, Beijing and Cangzllou, so did that
among Longdong, Guangzhou, wudi, Yuxian, Longzhong, Ohan, Huaiyin, Tianshui,
Neimengzhunger and Xinjiangdaye are more and more distance successively.
Neimengzhunger and Xinjiangdaye are the most distant from others.
References
Gao Zhenslleng et al .1994, Study on the comparison of Esterase Isozyme
between Creeping rooted and Non-creeping -rooted Alfalfa. Report of the
thirty-fourth North American Alfalfa Improvement Conference. Guelph . Canada
. 142.
GENETIC DIVERSITY IN WILD ACCESSIONS OF
MEDICAGO SATIVA FROM SPAIN.
E. Jenczewski, G. Genier, J. Ronfort and l.M. Prosperi.
INRA- Station de Genetique el Amelioration des Plantes, F-34130 Mauguio.
France
Rearing is a traditional component of the Mediterranean agriculture
system. Nowadays, it also represents the best way to valorize population-drained
marginal areas for agricultural, landscape maintenance and spare-time purposes.
Improvement programs thus require to focus on hardy and grazing-adapted
fodder types; pre requisites are firstly the collection and evaluation
of wild ecotypes diversity.
One hundred and four natural populations of Medicago sativa (called "Mielgas")
were collected in Spain, from September 1985 to July 1987, with the collaboration
of I. Delgado Enguita (SIA-DGA, Zaragoza, Spain). They were mainly from
roadsides, unirrigated and often grazed rangelands, or even orchards (olive
trees, vineyards...).
This germplasm has been evaluated at Montpellier, since 1986, under both
grazing and cutting managements. Most of the <> plants have
a prostrated habit and .l great ability of soil colonization (rhizomes).
Assessing the agronomic variability of the sampled populations notably
prove that this germplasm displays an important variability in growth rhythm
(the first cut in second year represents from 39% to 75% of the total annual
dry matter production, which itself accounts for 60% of the standard cultivars
production), a greater perenniality than alfalfa cultivars and a good vegetative
spring yield and seed production level for non-selected populations. Multivariable
analysis (figure 1) also demonstrate that the populations from the North-East
of Spain can be easily distinguished from the other "Mielgas",
since they behave as cultivated types.

Genetic diversity was also implemented through the survey of sixteen populations
with thirty RAPD markers. These populations were chosen in order to be
representative of the diversity which arose from the former analysis. The
distribution of RAPD polymorphism was assessed by partitioning this diversity
within and among populations components. This study confirmed the remarkable
intra-population diversity of <<Mielgas>>; indeed, in a previous
study concerning two populations (wild population/spanish cultivar), thirteen
primers uncovered sixty five fully reproducible fragments. Most of them
were polymorphic (93%) and indicated that 95 per cent of the variation
they displayed was partitionated within each population.
References
Delgado Enguita I., 1989. Estudio de la Variabilidad de las Mielgas
Aragonesas en areas de precipitation anual inferior a 600 mm. Tesis Doctoral-Universidad
Polytechnica de Madrid, Spain
Prosperi J. M., Delgado Enguita I., Angevain M., 1989: Prospection du
genre Medicago en Espagne et au Portugal. Plant Genetic Resources Newsletter
78/79, 27-29.
Prosperi J.M., Angevain M., Mansat P., 1990: Valorization of forage
genetic resources for selection of adapted cultivars. Example of wild mediterranean
lucernes. 95-98. in 6th Meeting of the FAO European sub-network on Mediterranean
Pastures and Fodder Crops. October 17/19, 1990. Bari (Italy).
Effect of Grazinq Frequency and Intensity
on Yield and
Persistence of Alfalfa-Tall Fescue Pastures.
Nestor A. ROMERO and N. Tony JUAN
Anguil Experiment Station, INTA
La Pampa (6326), Argentina.
There are five millon ha of alfalfa in the Pampean Region of Argentina.
In about 75 % of this area alfalfa is in mixture with perennial grasses,
mostly tall fescue (Festuca arundinacea L.). The use of moderately dormant
to nondormant alfalfa cultivars has increased since the early 80s. These
cultivars regrow faster after cutting or grazing, have higher forage production
in early spring and late fall, reach maturity faster, and have different
plant structure than dormant alfalfas. Differences among alfalfa genotypes
could affect alfalfa-grass competition and generate differential responses
to a given planting and grazing system. The objective of this study was
to evaluate the effect of alfalfa fall dormancy, row distance, and grazing
system on yield and persistence of alfalfa-tall fescue mixtures.
Twelve treatments were evaluated in a 2x3x2 factorial experiment, factors
were arranged in split blocks with six 1.6-ha paddocks as blocks. Factors
were: 1) Grazing system (GR1, 3,5 days of grazing + 17,5 days of resting;
stocking rate adjusted to graze tall fescue to half its height; and GR2,
7 days of grazing + 35 days of resting; stocking rate adjusted to leave
a 5-cm pasture residue); 2) Alfalfa fall dormancy (NDOR, nondormant cultivar
WL 605, FGS=9; MDOR, moderately dormant I cultivar WL 320, FGS=4; and DOR,
dormant II ecotype Pampeana, FGS=3); 3) Row distance (ROW15, 15 cm between
alfalfa and tall fescue alternate rows; and ROW30, 30 cm). Forage yield
(Mg DM ha-l) was estimated at the beginning of each grazing period during
three growing seasons. Persistence (ground cover as a % of initial cover)
was measured once a year.
Alfalfa yield was increased and tall fescue yield was decreased under the
GR2 grazing system as compared with GR1. With frequent but not intense
grazing (GR1) both alfalfa and fescue yielded more in ROW15 than in ROW30.
With less frequent but more intense grazing (GR2), alfalfa also yielded
more in ROW15, but the opposite happened with tall fescue. Yield of DOR
alfalfa was lower than yield of MDOR and NDOR alfalfa with both row distances.
Within ROW15, tall fescue yielded more associated with dormant alfalfa
than with less dormant ones. Persistence of tall fescue was higher under
GR1 than under GR2, regardless of alfalfa fall dormancy. All alfalfa genotypes
had higher persistence under GR2 than under GR1. Persistence of DOR alfalfa
was always lower than that of MDOR and NDOR alfalfa.
We conclude that, for our conditions, total yield of alfalfa-fescue mixtures
was affected by alfalfa dormancy and row distance, but not by grazing system.
Yield and persistence of alfalfa was increased when a moderately dormant
to nondormant genotype was planted at 15 cm between rows and managed with
a low frequency/high intensity grazing system. Yield and persistence of
tall fescue was highest when it was planted with a dormant alfalfa at 30
cm between rows, and grazed frequently but not intensively.
INBREEDING AS A METHOD OF DEVELOPING HOMOZYGOUS
LINES IN
ALFALFA, MEDICAGO SATIVA
E. V. Kvasova
Institute of Cytology and Genetics, Siberian Branch of the Russian Academy
of Sciences,
630090, Novosibirsk, 90, Russia
Applied genetics of self-incompatible species, such as alfalfa, has
been insufficiently developed because of the complex genetics nature of
their populations. It is known that some problems of applied genetics and
breeding can be solved by using homozygous lines. With this in mind we
produce homozygous lines by the method of inbreeding.
The results obtained with artificial population showed that alfalfa
inbred lines can be obtained only when a great number of plants from populations
are grown and analyzed. Thus, the total number of alfalfa depressed plants
in the first inbred generation was 83.4%. Plant vigor started to recover
after the third inbred generation. All the characters differentiate in
the first 2-3 generations. The differences in morphological characters
were wider among plants within than between lines. The widest phenotypical
variations in shape and size of leaf, flower and raceme, stem structure
were noted in the third generation. There were also variations in the size
and shape of pollen grains. The observed differences reduced with each
successive inbred generation so that the plants within the lines became
homogeneous by the 7-8th generations and the differences between lines
became clear-cut.
Based on the obtained results, it is concluded that inbreeding allows
to identify forms with any desirable character in a population. In development
of inbred lines, the aim of the experiment must be clearly defined for
the start. A group of self-fertile plants in a population suffices when
self-fertile forms are to be obtained. The greatest possible part of the
population gene pool must be involved when more diverse inbred lines are
to be produced. This can be achieved by a strategy overcoming the effect
of self-incompatibility system. This is because the group of selling plants
from which the inbred lines will be derived is too small for involving
the population gene pool.
WEEDALF$: An Integrated Expert System for
Weed Management in Alfalfa
J. F. Stritzke, C. W. Cuperus, and S. Zhang
Department of Agronomy and Entomology, Oklahoma State University
Stillwater, Oklahoma, 74078, U.S.A.
WEEDALF, the first version developed in 1989 dealt with the cost and
benefits of using herbicides in established alfalfa. The new version, WEEDALF$
(Version 2.0) has been upgraded to include an integrated approach to weed
management in both seedling and established stands of alfalfa. The four
main tasks of the system are weed management, weed information, herbicide
information, and profitability of using herbicides. Each component can
be accessed individually, but all are interconnected and can be utilized
in other sections.
With the "weed management" section, there is much accumulation
and integration of information to determine the weed problems and then
there is a listing of management alternatives. For example, with seedling
stands planted in the fall, integrated knowledge involves previous crop,
weed problems in previous crop, weed control used in previous crop, type
of seeded preparation, soil pH and fertility level, and type and extent
of weed problems. When weed control by herbicides is advised, the user
has the option of selecting from a listing of available labeled herbicides.
At this point in the program, the system switches to the herbicide information
section and allows the user to view general description and comments on
the various herbicides. Selection of the various herbicide options will
result in a cost/benefit display and an estimated profit with each option.
The "weed information" section contains knowledge about weeds
such as type, characteristics, and control options. The "herbicide
information" section contains description of herbicides currently
labeled on alfalfa. Information on each herbicide includes common name,
trade name, labeled use rate, current suggested retail cost, alfalfa growth
stage approved for application, growth stage of weed when effective, and
some general comments dealing with its application and effectiveness. The
"profitability of using herbicide" section is designed to calculate
profitability of a herbicide treatment compared to an untreated check.
The software was developed as a part of Oklahoma State's ongoing Integrated
pest Management program. The system is designed by using the PAD-BASED
model based on prototypes and delegation mechanism concepts, and developed
in the KnowledgePro(R) environment. The system requires a minimum of 640K
of RAM with DOS 3.0 or later version. A color monitor is recommended but
not required.
Breeding a Bloat-Tolerant Alfalfa in Argentina
D. H. Basigalup, C. V. Castell and C. D. Giaveno
EEA Manfredi/EEA Anguil - I.N.T.A. - Argentina
Partially financed by PRODUSEM, Pergamino, Argentina
With nearly 6.5 million ha, Argentina is the second largest alfalfa
producer in the world. Approximately 90% of this area is devoted to beef
and dairy production under direct grazing; therefore, bloat becomes a very
serious problem every year. An economical solution would be to have bloat-tolerant
alfalfa cultivars adapted to Argentine conditions. In 1991, the National
Institute for Agricultural Technology (INTA) initiated a breeding program
to develop such cultivars. In this paper we will briefly present that program
and discuss the progress obtained up to now.
The breeding goal is to select non-dormant cultivars with a significant
reduction in bloat potential. The objectives are: 1. To reduce the initial
rate of ruminal disappearance (IRRD); and 2. To maintain a good forage
quality level in the lower IRRD population.
Using the modified nylon bag technique (1), the program combines phenotypic
and genotypic selection (polycross progeny test) for reduced IRRD after
4 h in the rumen of fistulated steers. In 1995, seed of the lst cycle of
selection (C-l) was produced in pollination cages. Data from the progeny
test were also used to estimate IRRD broad sense heritability (h ) based
on variance component analysis.
Approximately 1300 C-l plants wer& transplanted to the field in September
1995 to initiate the 2n cycle of selection. Other 60 C-l plants were contiguously
transplanted to 60 C-0 plants in order to estimate selection progress on
IRRD reduction and to compare forage quality based on FRRD (= final rate
of ruminal disappearance after 72 h in the rumen), L:S (= leaf:stem ratio),
CP (= crude protein), ADF (= acid detergent fiber), NDF (= neutral detergent
fiber); and RFV (= relative feed value). The expected response to selection
(R) was estimated based on the equation: R = h . S, where h i s the heritability
for IRRD and selection differential.
Calculated mean h2 was 0.18 and S was 8.74%; therefore, R was estimated
to be 1.58%. The realized response to selection, based on the comparisons
between C-l and C-0 populations, was a reduction of 1.98% in IRRD. C-l
plants exhibited a significant higher fiber content than C-0 plants, but
this increase is still away from any detrimental effect on forage quality.
There was as much variability for all traits in the C-l population as in
the C-0 population, indicating possibilities for improving any trait if
necessary.
Reference
1. HOWARTH, R.E., B.P. GOPLEN, S.A. BRANDT, and K.-J. CHENG. 1982. Disruption
of leaf tissues by rumen microorganisms: An approach to breeding bloat-safe
forage legumes. Crop Sci. 22:564 568.&1
Genetic Variation for Digestibility and Fiber
Contents in the Medicago sativa Complex
B. Julier and C. Huyghe
INRA, Station d'Amelioration des Plantes Fourrageres, 86600 Lusignan, France
Alfalfa energetic value is limited by its low digestibility. Genetic
progress is necessary to increase or maintain the use of alfalfa forage
in ruminant feeding under intensive conditions. Breeding for quality implies
genetic variation for digestibility and related traits such as fiber contents,
and a breeding criterion. Improvement of forage digestibility must be done
without decrease in forage yield potential.
Digestibility and fiber contents were analyzed on 25 alfalfa populations
of the Medicago sativa complex. Populations from ssp. sativa and ssp. falcata,
diploid or tetraploid, were used. These populations proved to cover a large
range of variation for morphological traits (2). They were sown at Lusignan
(France) in a 2-rows plot design with 4 blocks in 1993. First cuttings
in spring in 1994 and 1995 were harvested. Forage yield was determined,
and stems were separated in 1994. Samples (stems in 1994, forage in 1995)
were dried, ground and analyzed for enzymatic digestibility and fiber contents
(NDF, ADF and ADL). Digestibility of NDF and lignin content in the cell
wall were calculated.
Analysis of variance were performed on the traits, and coefficients of
correlation calculated to determine tlle traits useful in breeding, i.e.
traits that were correlated with digestibility but not correlated with
forage yield. Genetic variation for digestibility was assessed? taking
into account the forage yield. Analysis of variance was performed on the
residues of the regression between digestibility and forage yield.
Wide genetic variation exits for these quality traits, at tlle sub-species
level and at the population level. Fiber contents (NDF, ADF and ADL) were
closely linked to digestibility but digestibility was negatively correlated
with forage yield. Digestibility of the cell wall and cell wall lignin
were correlated with digestibility and independent of forage yield. They
could be valuable breeding criteria to improve alfalfa digestibility without
reduction in forage yield. On average, at a given level of forage yield,
populations of the falcata sub-species were more digestible and had lower
fiber contents than populations of the sativa sub-species. Improvement
of cultivated alfalfa could involve crossings witl1 tl1e wild subspecies
falcata. These results confirm those already published (3). However, other
authors (1, 4) observed lower digestibilities in ssp. falcata related populations
than in ssp. sativa populations.
References
1. Buxton D.R., Hornstein J.S., Martin- G.C. (1987). Genetic variation
for forage quality of alfalfa stems. Can. J. Plant. Sci. 67, 1057-1067.
2. Julier B., Porcheron A., Ecalle C., Guy P. (l995). Genetic variability
for morphology, growth and forage yield
among perennial diploid and tetraploid lucerne populations (Medicago sativa
L.). Agronomie 15, 295-304.
3. Julier B., Guy P., Castillo-Acuna C., Caubel G., Ecalle C., Esquibet
M., Furstoss V., Huyghe C., Lavaud C.,
Porcheron A., Pracros r., Raynal G. (1996). Genetic vvariation for disease
and nematode resistance and forage quality
in perennial diploid and tetraploid lucerne populations (Medicago sativa
L.). Euphytica, in press.
4. Lenssen A. W., Sorensen E. L., Rosler G. L., Herbers L. H. (1991). Basic
alfalfa germplasms differ in nutritive content of forage. Crop Sci. 31,
293-296.
Biotechnological Improvement of Alfalfa
Nutritive Quality by Anti-sense Expression of
COMT and CCOMT. Methylating Enzymes in Lignin Biosynthesis
V. J. H. Sewalt, J. W. Blount, and R. A. Dixon
Lignin, a plant cell wall polymer derived from phenylpropanoid precursors
with varying numbers of aromatic methoxyl groups, interferes with forage
digestion to an extent that is dependent on its monomer composition (1).
Our objective is to improve alfalfa nutritive quality by reducing lignin
concentration and manipulation of extent of lignin methoxylation. We are
also working towards elucidation of the relative importance of different
methylating enzymes (O-methyltransferases) in monolignol biosynthesis.
Experiments were inititiated to manipulate lignin in alfalfa by downregulation
of COMT (caffeic acid O methyltransferase) and CCOMT (caffeoyl-CoA O-methyltransferase)
using anti-sense technology. A number of single and double anti-sense constructs
with COMT and/or CCOMT under control of the dual 35S promoter were introduced
into alfalfa by Agrobacterium-mediated transformation. Controls were untransformed,
but regenerated plants, and plants harboring 35S-GUS (pBI121 plasmid).
The two controls were not different in OMT activities or lignin characteristics.
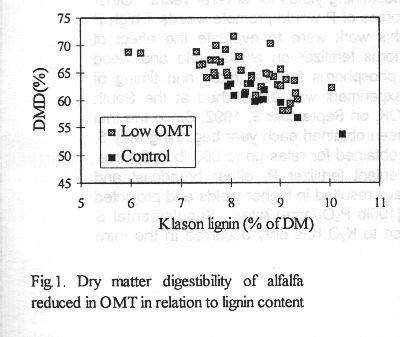
A number of plants were obtained in which COMT, CCOMT, or both OMTs
were reduced. Most antisense plants with reduced OMT activity and all plants
harboring pBI121 were MPT II-positive by ELISA-assay. Down-regulation of
COMT and/or CCOMT in several primary transformants resulted in modestly
reduced lignin concentration and/or extent of methoxylation. One single
COMT antisense transformant was strongly reduced in lignin, and its lignin
displayed a sharp increase in methoxyl content, indicative of an increase
in S/G ratio. Interestingly, cell wall (NDF) concentration was drastically
reduced in many lines. Enzymatically determined digestibility of NDF improved
only slightly in many lines, but due to the reduction in NDF content, most
transgenic lines had significantly higher dry matter digestibility. Further
lignin analysis of promising lines and confirmation of transgene expression
and copy number is currently underway. The preliminary results of this
experiment demonstrate the feasibility to generate improved alfalfa lines
with reduced lignin and altered lignin composition for improvement of digestibility.
Reference
1. Sewalt, V. J. H., W. G. Glasser, J. P. Fontenot, and V. G. Allen.
1996. Lignin impact on fiber degradation. 1.
Quinone methide intermediates formed from lignin during in vitro fermentation
of corn stover J. Sci. Food
Agric. (in press).
ALFALFA YIELD RESPONSE TO METHOD AND RATE
OF APPLIED P AND
WEED INTENSITY
S.B. Phillips, G.V. Johnson, and W.R. Raun
Oklahoma State University, Stillwater, Oklahoma
ABSTRACT
Past experience in Oklahoma indicates that initial and maintained soil
fertility levels strongly affect long-term production of alfalfa (Medicago
sativa L.). Presently, there are indications which suggest that P fertilizers
should be applied at two to three times the recommended rate at the time
of establishment when the fertilizer can be incorporated. Preplant band
applied P fertilizer may provide increased long term benefit if soil-fertilizer
P reactions are reduced. High rates should provide a P fertility foundation
with the potential for sustaining yields for several years. Other work
suggests that carrier N in some sources of P would promote weeds if high
P rates were used. The objectives of this work were to evaluate the effect
of alternative methods of applying phosphorus fertilizer on alfalfa yield
and weed populations and to determine optimum phosphorus fertilizer rates
and timing of application for alfalfa production. The experiment was established
at the South Central Research Station in Chickasha, OK, on September 9,
1992. Since the trial was initiated, five forage harvests have been obtained
each year beginning May 5, 1993. A linear response to P has been obtained
for rates up to 600 Lb P205/acre. Single applications of high rates of
preplant fertilizer P, either broadcast and incorporated or deep knifed
in a band, have resulted in higher yields and projected higher profits
than annual conventional (100lb P205/acre) rates. Supplemental S has not
shown a response and response to K2O has only occurred in the third year.
Aluminum Tolerance QTL in Diploid Alfalfa
M.K. Sledge, J.H. Bouton, J. Tamulonis, G. Kochert, and W.A. Parrot
Department of Crop and Soil Sciences , and Department of Botany,
University of Georgia,
Athens, Georgia, 30602 USA
Aluminum toxicity is the major growth-limiting factor for crop production
on acid soils, which comprise up to 40% of the world's arable lands, including
much of the United States. Surface liming, used to correct soil acidity,
is expensive and does not neutralize harmful levels of subsurface aluminum.
Alfalfa has been shown to be sensitive to aluminum toxicity. but conventional
breeding methods have made little progress in introducing aluminum tolerance.
We report the identification of quantitative trait loci (QTL) controlling
aluminum tolerance in diploid alfalfa. Probes from our existing alfalfa
RFLP map were used to identify and map the QTL. An in vitro callus growth
bioassay was used to select aluminum tolerant and aluminum sensitive genotypes.
The aluminum sensitive ssp. coerulea genotype (440501-2) was crossed with
an aluminum tolerant genotype from a ssp. coerulea PI (464724) to produce
and F2 population. The parents were screened with 146 cDNA probes, and
58 were mapped to nine linkage groups. The F2 genotypic classes for each
of the 58 RFLP loci were contrasted with means from the callus growth bioassay,
using ANOVA. We also used Mapmaker-QTL to identify markers associated with
aluminum tolerance. Four markers, UGAc044. UCAc053, UGAc141, and UGAc782,
were found to be associated with aluminum tolerance. UCAc044 had the greatest
effect, accounting for 15% (LOD 2.3) of the variation in aluminum tolerance.
These markers will aid in the future introgression of aluminum tolerance
from tolerant germplasm into cultivated alfalfa. We are currently working
to isolate allele specific markers near the UGAc044 locus, using bulked
segregant analysis and AFLPs. Markers specific for the aluminum tolerant
allele can be converted to sequenced tag sites and used to quickly screen
large numbers of plants for the tolerant allele. We have isolated 5 bands
specific for the tolerant allele, and 7 bands that are specific for the
sensitive allele.
Brummer, E. C., J. H. Bouton, and G. Kochert. 1993. Development of an
RFLP map in diploid alfalfa. Theor. Appl. Cenet. 86:329-332.
Michelmore, R. W., T. Paran, and R. V. Kesseli. 1991. Identification of
markers linked to disease-resistance genes by bulked segregant analysis:
A rapid method to detect markers in specific genomic regions by using segregating
populations. Proc. Natl. Acad. Sci. USA 88:9828-9832.
Parrot, W. A., and J. H. Bouton. 1990. Aluminum tolerance in alfalfa as
expressed in tissue culture. Crop Sci. 30:387-389.
Lincoln, S. E. M. J. Daly, and E. S. Lander. 1993. Mapping genes controlling
quantitative traits using MAPMAKER/QTL version 1.1: A tutorial and reference
manual. Whitehead Institute, Cambridge, Mass.
Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A.
Frijters, J. Pot, J.Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new
technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414.
Predicting Acid/Aluminum Tolerance In
Alfalfa Genotypes with Hematoxylin Staining.
Kelley, Rowena Y., Powell, D.; Yang, G.; Glass, M. North Carolina A&T
State University, Greensboro , NC., 27411
Alfalfa (Medicago sativa L.), one of the most important forage legumes
in the United Stales,
has been recognized as an aluminum sensitive species . Hematoxylin staining
pattern is a technique that
has been used to evaluate differences in root growth and stain uptake between
wheat cultivars sensitive
or resistant to high aluminum concentrations. Thc hematoxylin staining
pattern was the focus of this
study because the procedure is simple, rapid and uses minimal space and
labor. The objectives of this
study were: 1. To determine if hematoxylin staining can be used for evaluating
acid/aluminum tolerance
in alfalfa 2. To determine the proportion of aluminum tolerant individuals
for various alfalfa cultivars
with use of hematoxylin staining. Ten different alfalfa cultivars werc
used: Apollo, ARC, Foundation
Vernal, Shenandoah, Spreador 2, WL 311, Saranac, Saranac AR, Cimarron,
and Cimarron VR Forty
seeds from each cultivar were planted in Metro-mix 360 for use as controls.
Forty seeds of each type
were also stained in a solution of hematoxylin for a period of two days.
After staining, the seedlings
were transferred to Metro-Mix 360 for twenty-one days. After twenty-one
days, the plantlets were
transferred to Porter's soil, an 80% aluminum saturated soil with a pH
of 4.5, and grown in the
greenhouse for sixty days. After sixty days, fresh and dry root and shoot
weights were determined and
performance, based on biomass, was compared to the tolerance level predicted
by hematoxylin stain.
This study showed that some individuals in each cultivar were tolerant
to the high concentrations of
aluminum (80% saturation). Individuals which scored >2 showed similar
results with the most
significant difference being the numbers of individuals found at the various
stain levels.
Cis-elements and Trans-acting Factors Required
for
the Regulation of Alfalfa Isoflavone Reductase
B. Miao and N. Paiva
Plant Biology Division,
The Samuel Roberts Noble Foundation, Inc.
P. O. Box 2180, Ardmore, OK 73402
We are using both plant cell cultures and transgenic plants to study
the regulation of isoflavonoid phytoalexin biosynthesis. In the isoflavonoid
biosynthetic pathway, isoflavone reductase (IFR) is specific for the synthesis
of medicarpin, the major antifungal isoflavonoid phytoalexin produced by
alfalfa in response to fungal pathogen attack. Low levels of IFR mRNA and
medicarpin conjugates accumulate constitutively in alfalfa cell cultures
and root system. However, following fungal elicitor addition, high levels
of medicarpin accumulate following rapid transcriptional activation of
the IFR gene. Organ-specific expression may also become a strong local
response when the plant is subjected to the infection. To understand how
the IFR gene is regulated, the IFR promoter has been analyzed both in vitro
and in vivo .
By using promoter fragments in gel retardation assays (GRAs), a specific
DNA-binding complex was observed by using nuclear extracts from both elicited
and unelicited cell cultures. The level of binding activity increases about
5-fold within 90 min of elicitation. At later time points, the signal decreased
rapidly, indicating the response to elicitation is rapid but transient.
Further detailed GRA analysis both indicated that an AT-rich region from
-270 to -130 bp upstream of the start of transcription is essential for
transcription factor binding. In this same AT-rich region, DNase I footprinting
revealed two elicitor induced regions, Box I and Box II. Both sites are
about 20 bp long and AT-rich with very similar DNA sequences. The functions
of these elements were tested in transgenic alfalfa plants using a series
of IFR promoter deletion-GUS fusion constructs. Deletion of Box I and Box
II appears to greatly reduce the expression of the GUS reporter gene in
vivo. By using nuclear extracts, the protein blots were probed with either
IFR fragment B or the synthetic multimer including consensus sequence of
Box I and Box II. The Southwestern blot analysis revealed a 50-60 kD protein
band. Phosphatase treatment eliminates the DNA binding activity of nuclear
extracts, suggesting that the factor may be present at all times, then
activated via phosphorylation by a protein kinase element of a signal transduction
pathway.
References 1. Oommen, A., Dixon, R. A., Paiva, N. L., 1994. The elicitor-inducible
alfalfa isoflavone reductase promoter confers different patterns of developmental
expression in homologous and heterologous transgenic plants. Plant Cell
6: 1789-1803.
Paiva, N. L., Edwards, Y. Sun, Y., Hrazdina, G., Dixon, R. A., 1991.
Stress responses in alfalfa (Medicago sativa L.). 11. Molecular cloning
and expression of alfalfa isoflavone reductase, a key enzyme of isoflavonoid
phytoalexin biosynthesis. Plant Mol. Biol. 17: 653 -667.
Toward Metabolic Engineering of Alfalfa for Production
of Prenylated
Isoflavonoid/Pterocarpan Phytoalexins
Z. Guo and N. L. Paiva
Plant Biology Division, The Samuel Roberts Noble Foundation,
2510 Sam Noble Parkway, Ardmore, Oklahoma 73402, U. S. A.
Upon pathogen infection, alfalfa (Medicago sativa L. ) accumulates the
simple pterocarpan phytoalexin medicarpin as a defense response. Some legumes,
such as beans (Phaseolus vulgaris), soybean ( Glycine max), and white lupin
(Lupinus albus), produce prenylated isoflavonoids and/or pterocarpans either
upon elicitation or in a constitutive manner (1). In contrast, no prenylated
isoflavonoids have been reported to accumulate in alfalfa. It has been
shown that prenylated isoflavonoids and pterocarpans are more fungitoxic
than their non-prenylated counterparts. Also, prenylated isoflavonoids
were shown to inhibit feeding of insect herbivores. Aiming at improving
fungal and pest resistance, we are exploring the feasibility of altering
the phytoalexin profile in alfalfa by introducing a suitable exotic prenyltransferase.
We focused on the bean system as a source because of the high structural
similarity between its prenylation substrate and medicarpin and because
of a previous report that bean prenyltransferase is active on medicarpin
(2).
Using a microsomal preparation from CuCl2-treated bean as catalyst, we
demonstrated stereospecific biosynthesis of 10-dimethylanylmedicarpin from
dimethylanyl diphosphate and (-)-medicarpin. Structural verification of
the reaction product was achieved using chromatographic and W, MS and lH-NMR
spectral analyses. We are currently performing bioassays to determine the
fungitoxicity of this product relative to medicarpin. Medicarpin prenylation
activity appeared 4-8 hours after CuC12 induction; it increased and reached
the maximum level in about 32 hours. The accumulation of the phytoalexins
phaseollin and kievitone followed a similar pattern. Earlier reports indicated
that the pterocarpan prenyltransferase is associated with chloroplast membranes,
and so we developed a protocol for the large scale preparation of chloroplast
envelope membranes (CEM) from bean seedlings that are treated with CuCl2
to induce high levels of prenyltransferase activity. We were able to show
that this preparation had specific prenylation activity 150 fold higher
in comparison with microsomal fractions. Further purification (double the
specific activity) was achieved by differential solubilization. Rabbit
antisera against partially purified CEM proteins, both native and denatured
were generated and used to screen a bean cDNA expression library. A total
of 144 positive clones have been isolated. Work is underway to characterize
these clones according to CuCl2-inducibility and sequence similarity to
known prenyltransferases.
References
1. Biggs, D. R., Welle, R., and Grisebach, H. 1990. Intracellular localization
of prenyltransferases of isoflavonoid phytoalexin biosynthesis in bean
and soybean. Planta 188:244-248.
2. Biggs, D. R., Welle, R., Visser, R. F., and Grisebach, H. 1987.
Dimethylallylpyrophosphate:3,9-dihydroxypterocarpan-10-dimethylallyl
transferase from Phaseolus vulgaris. FEBS Letters 220:223-226.
Alfagene.s - A Comprehensive Internet-accessible
ACE Database for Alfalfa
http://probe.nalusda.gov
D. Z. Skinnerl,2, P. C. St. Amand1,2, and S. M. Ramaiah2
USDA-ARS and Agronomy Department, Kansas State University
Manhattan, Kansas, 66506-5501, U.S.A.
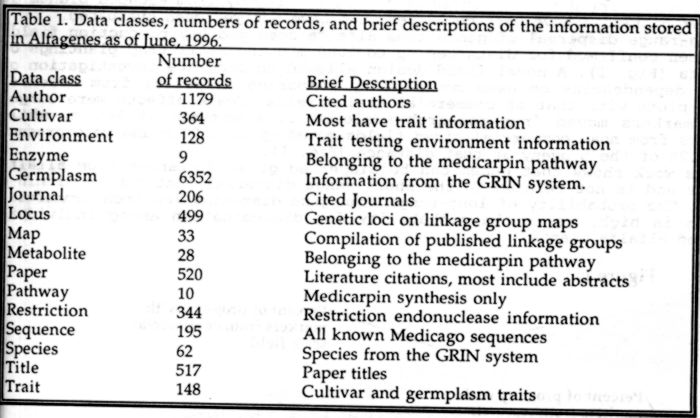
A comprehensive computer database of the alfalfa genome has been created
using the powerful "ACE" (1) software. The goal of this database
is to make all known information relevant to the alfalfa genome accessible
from a single location. Currently, the database contains information in
17 classes (Table 1). Data have been compiled from numerous sources, including
the USDA Germplasm Resources Information Network (GRIN), The Certified
Alfalfa Seed Council, and hundreds of published works. The data are inter-related
such that information in one data class can lead immediately to information
in another class simply by following a hypertext link. For example, a DNA
sequence that is cited in a literature citation can be displayed immediately
simply by selecting the name of the sequence from within the literature
citation. Numerous powerful search and analysis tools are available within
the database. For example, DNA sequences may be fully analyzed with resident
tools that can generate restriction endonuclease maps, or search for open
reading frames. Graphic images of linkage groups can be displayed, and
recombination frequencies are available. Graphic images of metabolic pathways
are available, selecting any part of the pathway displays complete information
on that part. Search tools can be applied to any part of the database;
for example, literature citations may be searched for keywords or specific
authors. The database is accessible on the world-wide-web at: http://probe.nalusda.gov
.
Reference
1. Richard Durbin and Jean Thierry Mieg (1991-). A C. elegans Database.
Documentation, code and
| data available from anonymous FTP servers at lirmm.lirmm.fr, cele.mrc-lmb.cam.ac.uk
and
ncbi.nlm.nih.gov.
Risk of Alfalfa Transgene Dissemination and
Scale Dependent Effects
P. C. St. Amandl, D. Z. Skinner1, and R. N. Peaden2
lUSDA/ARS and Agronomy Department, Kansas State University, Manhattan KS
66506 and USDA/ARS, Irrigated Agricultural Research Center, Prosser WA
99350
Concerns have been raised over the release of genetically engineered
plants into the environment. Because pollen can readily disseminate engineered
genes throughout a plant population or into a related species, the risk
of inadvertent dispersal of engineered genes must be known. Measures of
that risk include the rate of pollen-mediated gene spread, the maximal
dispersion distance of pollen, and the spatial dynamics of pollen movement
within seed production fields; none of which are known for alfalfa (Medicago
sativa L.), ïan insect pollinated species. Lacking from most risk
assessment studies is the investigation of scale dependencies of the data.
Using a rare, naturally occurring molecular marker, pollen dispersal can
be tracked without introducing engineered genes into the environment. Using
PCR-based methods, a suitable marker was found in an intron of the alfalfa
glutamine synthetase gene. This marker was not present in a broad range
of alfalfa genotypes, and was used to measure gene dispersal within seed
production fields. Preliminary results indicate that bees (Megachile spp.)
used in commercial seed production show a bi-directional bias when pollinating,
resulting in the movement of pollen from marker plants directly toward
and away from the colony hive. Gene movement within fields from individual
plants occurs only over very short distances, up to 4 m in this study.
By examining widely-dispersed, individual escaped alfalfa plants and their
progeny using RAPD markers, gene movement among escaped alfalfa plants
has been confirmed for distances up to 230 m. Progeny from escaped plants
at greater distances have been collected, but have not yet been evaluated.
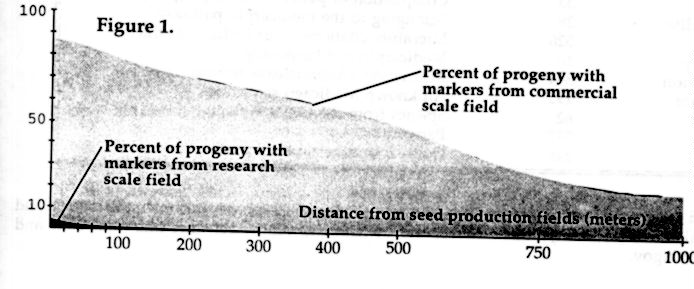
Long-range dispersal of genes from alfalfa seed and hay production fields
has been confirmed for distances up to 1000 m using small trap plantings
of alfalfa (Fig. 1). A novel field design allowed an accurate investigation
of scale dependencies on gene movement by comparing dispersal from research
scale plots with that of commercial scale fields. Scale effects were large.
Gene markers moved from research scale plots a maximum of 300 m, while
markers from near commercial sized fields moved up to 1000 m and were present
in 22.2% of the progeny at that distance (Fig. 1). This work shows that
genes can be dispersed great distances from alfalfa fields and is not related
to the short range dispersal that occurs within a field. The probability
of long-range transgene dissemination from commercial fields is high, as
is the probability for dissemination among individual escaped alfalfa plants.
Mapping of Simple Sequence Repeats (SSR) DNA
Markers in Diploid
and Tetraploid Alfalfa
Noa Diwan1,2, Joseph H. Bouton3, Gary Kochert4, Arvind A.
Bhagwat1,2, and Perry B. Cregan1.
lSoybean and Alfalfa Research Lab., USDA-ARS, Beltsville, MD.
20705. Department of Agronomy, University of Maryland, College
Park, MD 20742. 3 Department of Agronomy, University of Georgia,
Athens, GA 30602. Department of Botany University of Georgia,
Athens, GA 30602.
Cultivated alfalfa (Medicago sativa) is an autotetraploid. However,
all three existing alfalfa genetic maps resulted from crossing of diploid
alfalfa. A study was undertaken to evaluate the use of Simple Sequence
Repeat (SSR) for mapping diploid and tetraploid alfalfa. Ten SSR loci were
incorporated into an existing F2 diploid alfalfa RFLP map, and were also
mapped in an F2 tetraploid population. The tetraploid population had two
to four alleles in each of the loci examined. Following the segregation
of these alleles in the tetraploid mapping population generally was clear
and easy. Linkage relationships at the tetraploid level were determined
by using a single dose allele
(SDA) analysis, where each allele is scored as a dominant marker independently
of the other alleles at the same locus. The SDA diploid map was also constructed
to compare mapping using SDA to the standard co-dominant method. Linkage
groups were generally conserved among the tetraploid and the two diploid
linkage maps, except for segments where severe segregation distortion was
present. Segregation distortion which was present in both tetraploid and
diploid populations probably resulted from
inbreeding depression. Therefore, a better approach to map
alfalfa should be in a non-inbred mapping population such as F1.
The ease of analysis together with the abundance of SSR loci in
the alfalfa genome indicated that SSR markers should be a useful
tool for mapping in tetraploid alfalfa.
References
Brummer, E.C., J.H. Bouton, and G. Korchet. 1993. Development of
an RFLP map in diploid alfalfa. Theor Appl. Genet. 86:329
332.
Cregan, P.B., A.A. Bhagwat, M.S. Akkaya, and Jiang Rongwen.
1994. Microsatellite fingerprinting and mapping of soybean.
Methods. Mol. Cell. Biol. 5:49-61.
D. A. Silva, J. A. G., M. E. Sorrells, W. L. Burnquist, and S. D.
Tanksley. 1993. RFLP linkage map and genome analysis of
Saccharum spontaneum. Genome, 36:782-791
Yu, K. F., and K. P. Pauls. 1993. Segregation of random amplified
polymorphic DNA markers and strategies for molecular mapping
in tetraploid alfalfa. Genome, 36:844-851.
Divergent Selection for Self-fertility in
Alfalfa and Correlated Responses for Forage Yield
D. Rosellini, F. Veronesi and G. Barcaccia
Istituto di Miglioramento Genetico Vegetale, University of Perugia
Borgo XX Giugno 74, 06100 - Perugia, Italy
Different breeding strategies are being followed in advanced alfalfa
breeding. The role of recurrent selection in population improvement has
been recently reexamined in the light of the greater importance attributed
to 'complementary gene interactions' with respect to allelic interaction
in determining vigor. According to this interpretation, inbreeding in the
selection process could help reduce the genetic load and fix favorable
linkage groups. A different breeding strategy aims at developing double
cross cultivars based on well combining, self-sterile clones through the
somatic seed technology. Self-fertility in alfalfa has been widely studied:
it shows continuous variation in the populations, is associated to cross-fertility
and is drastically reduced by inbreeding, usually more than forage yield.
Even if a gametophytic self-incompatibility system has been suggested,
the cause of self sterility seems to be a 'relational incompatibility'
that can be related to genetic load. If the genetic loads for fertility
and for forage yield are at least partly coincident, selection for self-fertility
may help reduce the genetic load and bring about positive correlated responses
for forage yield. In this perspective, selecting SS genotypes for developing
hybrids would not be a valuable strategy. Divergent selection for self-fertility
was applied in a central Italian alfalfa landrace with the purpose of developing
materials to test the previous hypotheses. Four hundred and fifty three
plants were grown in pots under bee proof cages and evaluated for self-fertility
(seeds produced per floret tripped, 30 to 40 florets per plant); 10 self-fertile
(SF) and 10 self-sterile (SS) plants were selected on the basis of 3 evaluations
of self-fertility made in different environmental conditions. The 10 clones
of each group were hand crossed without emasculation according to a n(n-l)
diallelic design; equal amounts of seeds from reciprocal crosses were pooled,
thus obtaining 45 full-sib families per group. Ten plants per family were
grown in pots randomized within bee-proof cages and evaluated for self-fertility.
The following spring, 5 random plants per family were evaluated for green
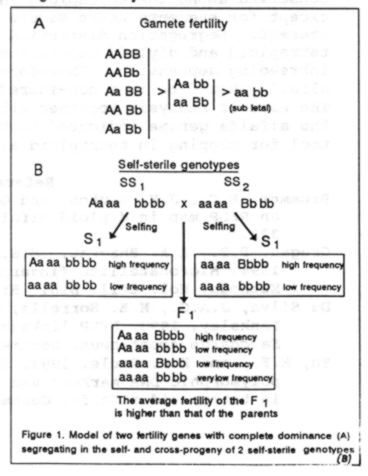
matter yield. Selection was effective for self-fertility (h2R=0.52), but
not for self-sterility. Such results are expected if self-sterility is
due to genetic load (figure 1). The SF plants, on the contrary, would have
a low genetic load, i.e., a high frequency of dominant genes for fertility,
and their progenies would show improved self fertility. This hypothesis
is consistent with the low cross-fertility of the SS plants observed in
the diallel cross, when compared to that of the SF clones. The forage yield
of SS progenies (21.7 g per plant) was significantly higher than that of
SF progenies (13.6), which did not differ from the control (13.22). This
could be due to a lower than normal percentage of Sl plants in the SS progeny.
A dense stand trial is underway to verify whether the forage yield superiority
of SS progeny is mantained in competitive conditions, in which Sl plants
are selected against. Inbred SF materials are also being developed to test
the effect of deliberate selection for self-fertility during inbreeding
on vigor.
Ploidy Determination of Medicago sativa
ssp. falcata germplasm:
Step 1 in a falcata germplasm enhancement program
E. C. Brummer and S. Sin
Agronomy Department, Iowa State University
Ames, IA 50011 USA
Alfalfa, Medicago sativa, breeding programs in most of the United States
have traditionally concentrated on germplasm containing predominantly ssp.
sativa. M. sativa ssp. falcata has been used only to a limited extent,
mainly for northern and rangeland areas due to its generally superior winter-
and drought tolerance. For other traits such as forage yield and disease
resistance, falcata material is inferior to .sativa. Molecular studies
have shown that falcata germplasm is genetically distinct from sativa (Kidwell
et al. 1994, Brummer et al. 1991) indicating that it may include desirable
alleles for various agronomic traits. Bingham et al. (1995) contended that
alfalfa heterosis is due to the presence of complementary linkage blocks.
However, Barnes et al. (1977) pointed out that the current methods used
to breed alfalfa may be breaking apart desirable blocks, bringing genes
and alleles controlling important traits into equilibrium. Thus, the addition
of new linkage blocks may be necessary to allow continued improvement in
alfalfa cultivars and enhanced falcata germplasm is one possible source
of these blocks.
The long term objective of this project is to develop falcata germplasm
that may be useful for alfalfa breeding programs. The current germplasm
collection consists of mostly unimproved materials, which have not seen
several decades of concerted breeding efforts as has sativa. The specific
objective of this research is to determine the ploidy level of all available
falcata acccssions. We plan to use the tetraploid accessions in further
breeding work.
Ploidy has been determined on 159 falcata accessions to date. All materials
were obtained from the Western Regional Plant Introduction Station at Pullman,
WA. Chromosome counts were made on root tip squashes from young seedlings
grown in petri plates. Five plants per accession were counted; a minimum
of 10 cells per plant were observed. Approximately 63% of the accessions
counted were tetraploid, 33% diploid, and 6% mixtures of diploids and tetraploids.
Enhancing the falcata germplasm pool should be conducted at the tetraploid
level ir the material is intended to be introgressed into current alfalfa
breeding material. Thus, determination of the ploidy of each accession
is useful prior to planting selection nurseries. Actual chromosome counts
are the best way to assess ploidy because morphological differences are
not always reliable indicators. The fact that almost two-thirds of the
falcata accessions for which seed is available are tetraploids will allow
us a large diversity of material from which to begin selection.
Although the enhancement of this germplasm will not be easy or its results
quickly applied to current cultivar breeding programs, without this breeding
work little use will ever be made of the falcata collection. This falcata
enhancement project will use the identified tetraploid accessions as the
basis from which to select for various agronomic traits, including winterhardiness,
upright growth, leafiness, and later fall dormancy. Even though falcata
germplasm introgression into traditional hay cultivars may be limited,
this material may find a larger niche in cultivars developed specifically
for long-term permanent pastures, where intensive cutting and management
systems are of less importance. A shift in the paradigm of alfalfa breeding
may result in further use of enhanced falcata germplasm.
References:
Barnes, D.K., E.T. Bingham, R.P. Murphy, O.J. Hunt, D.F. Beard, W.H. Skrdla,
and L.R. Teuber. 1977.
Alfalfa germplasm in the United States: Genetic vulnerability, use, improvement,
and maintenance.
USDA Tech. Bull. No. 1571. USDA-ARS, Washington, D.C. 21 p. Bingham, E.
T., R. W. Groose, D. R. Woodfield, and K. K. Kidwell. 1994. Complementary
gene interactions in
alfalfa are greater in autotetraploids and diploids. Crop Sci. 34:823-829.
Brummer, E. C., G. Kochert, and J. H. Bouton. 1991. RFLP variation in diploid
and tetraploid alfalfa Theor.
Appl. Gen. 83:89-96. Kidwell, K. K., D. F. Austin, and T. C. Osborn. 1994.
RFLP evaluation of nine Medicago accessions
representing the original germplasm sources for North American alfalfa
cultivars. Crop Sci. 34:230-236.
Progeny Test Based on Morphological and
Molecular Markers to Verify the Occurrence of
Parthenogenesis in an Apomeiotic Mutant of Diploid Alfalfa
G. Barcaccia1, S. Tavoletti2, M. Falcinellil and F. Veronesi2
(1) Istituto di Miglioramento Genetico Vegetale, University of Perugia
(Italy);
(2) Dipartimento di Biotechnologie Agrarie ed Ambientali, University of
Ancona (Italy).
In the polysomic polyploids, such as cultivated alfalfa, maximum heterosis
may be expressed by a few elite individuals of the population but not by
the entire population. An approach to perpetuate the elite individuals
and preserve heterosis over generations could be the use of apomixis. The
apomictic reproduction consists of two essential processes: apomeiosis
(formation of eggs avoiding meiotic reduction) and parthenogenesis (development
of embryos without fertilization). Thus, apomixis has the potential of
cloning plants through the seed and it furnishes a unique opportunity for
developing superior tetraploid cultivars with permanently fixed heterosis.
Apomixis as a whole has not been detected in the genus Medicago, but components
of apomixis are present. The formation of unreduced eggs through apomeiosis
in a diploid plant of M. sativa subsp. falcata (L.) Arcang., named PG-F9
is an extremely interesting feature of apomixis, as is the induction of
haploid parthenogenesis in tetraploid lines of alfalfa, which has been
widely exploited to obtain CADL populations. Our goal is to eventually
combine all the components of functional apomixis in alfalfa. Studies conducted
in diplosporous species report that apomeiosis and parthenogenesis could
be processes tightly associated. Information on the genetic composition
of progenies from the mutant PG-F9 cannot be obtained on the basis of cytoembryological
analyses. Therefore, a progeny test based on morphological and molecular
markers was carried out to verify the parthenogenetic capability of apomeiotic
eggs. Four morphological traits such as leaf shape, stipule form, stem
pigmentation and flower color showed to be effective in the preliminary
screening of progenies and most of the plants were classified as aberrants
(from sexual reproduction). Molecular investigations by means of RAPD fingerprinting
and RFLP marker detection conducted in the progenies morphologically classified
as maternal allowed two plants genetically similar but no identical to
PG-F9 to be discovered. Owing to the high number of molecular markers conserved
as in PG-F9 and because the great discriminating efficiency of primers
and probes used, these plants could likely be generated through parthenogenesis
of apomeiotic eggs. According to the degree of similarity distribution
in the self progeny, the differences found at few genomic loci could be
explained by an origin from selfing only assuming a non-ordinary conservation
of the maternal genotype. In fact, considering the PG-F9 is not inbred,
there was a remote possibility to find a plant from selfing identical to
PG-F9 by using the 36 selected ten-mer primers (that generated in PG-F9
a total of 269 RAPD bands) and by investigating the 12 heterozygous RFLP
loci. The presence of DNA rearrangements could be explained by the occurrence
of FDR mechanisms, which were not observed at the cytological level, whereas
an autosegregation process should not be involved since the metaphase analysis
revealed the normal somatic chromosome number. The overall judgment about
the sexual or apomictic origin of the plants morphologically e molecularly
similar to the apomeiotic mutant PG-F9 will require further investigations
at the nucleotidic level.
Assessment of genetic variability for
cryoprotective oligosaccharide accumulation in cold acclimated leaves of
alfalfa.
Y. Castonguay, P. Nadeau & R. Michaud. Agriculture and Agri-Food Canada,
Soils and Crops Research and Development Centre, Sainte-Foy, Quebec, Canada,
GlV 2J3.
Cold acclimation of alfalfa is associated with marked changes in the carbohydrate
composition of overwintering crowns and roots (McKenzie et al., 1988).
We have recently shown that differences in freezing tolerance between winterhardy
and nonhardy cultivars (cvs.) are more closely related to crown levels
of the oligosaccharides of the raffinose family (RFO; stachyose and raffinose)
than to the levels of sucrose (Castonguay et al., 1995). This observation
suggests that cryoprotective RFO could constitute a potential trait to
screen for cold tolerance in alfalfa. However, the extent of genotypic
variation for RFO accumulation within populations of alfalfa and its potential
for the genetic improvement of cold tolerance of alfalfa needs to be further
investigated. The objectives of this study were to assess whether RFO accumulate
in leaves of alfalfa and to determine the extent of the variability of
leaf RFO within an alfalfa cv. This information is important in order to
develop a non-destructive method to screen alfalfa genotypes for their
potential to accumulate cryoprotective oligosaccharides.

We found that raffinose accumulates in cold-acclimated leaves whereas
stachyose remained at very low levels. Cold-induced accumulation of raffinose
gradually increases from non-hardy to very hardy cvs. Superior levels of
leaf raffinose in plants acclimated at a nonlethal subfreezing temperature
relative to plants acclimated at a low non-freezing temperature further
indicate that leaf raffinose is closely linked to freezing tolerance in
alfalfa. Analysis of leaf raffinose in 497 genotypes of the cv. DK-125
showed a wide distribution varying from nearly 0 up to 20 mg g-1 dry weight
raffinose with a mean skewed toward lower values (Fig. 1). We observed
a contrasting accumulation of leaf raffinose between cold acclimated cuttings
from genotypes selected either for their high or low level of leaf raffinose.
The extent of the variability within each genotype indicates that the variation
in cold-induced accumulation of leaf raffinose within a population is genetically
determined. Our results suggest that cold-induced accumulation of leaf
raffinose could potentially be used as a non-destructive screening method
to improve cold tolerance in alfalfa.
References
Castonguay, Y., Nadeau, P., Lechasseur, P. and L. Chouinard. 1995. Differential
accumulation of carbohydrates
in alfalfa cultivars of contrasting winterhardiness. Crop Science 35: 509-516.
McKenzie, J. S., R Paquin, and S. H. Duke. 1988. Cold and heat tolerance.
P. 259-302. In A. A. Hanson et al.
(ed.) Alfalfa and alfalfa improvement. Agron. Managr. 29. ASA, CSSA, and
SSSA, Madison, WI.
Analysis of Seed Storage Proteins in Regenerated
Plants of Alfalfa
(Medicago varia Mart.)
O. Tzeveleva., S. Peltek, E. Deineko
Institute of Cytology and Genetics, Novosibirsk, Russia
Different biotechnological approaches can be utilized for inducing more
variable cross pollinated species (for instance, tetraploid alfalfa). One
of them is based upon the phenomenon of somaclonal variation. Alfalfa somaclones
<Ire widely used as a source of variation for basic research and breeding.
It is often impossible to confirm the genetic nature of this phenomenon.
To understand the origin of somaclonal variation, we found it most appropriate
to use seed storage proteins, as a part of them are largely under monogenic
control, they are easy to identify, and allow a large number of phenotypes
to be analyzed at a time. The experimental material on which we studying
changes in the pattern of seed storage proteins in alfalfa regenerants
(Medicago varia Mart.) was obtained as indicated in Fig.1. The patterns
were analyzed on the self-pollinated progeny of all the three donor regenerant
pairs after SDS- PAAG. About 100 seeds of each of the plants were analyzed
individually. In total, 38 individual components have been revealed. We
counted the number at which each of the 38 components occurred in the patterns.
If each component were under monogenic control, the expected frequencies
would be as follows: AAAA-1; AAAa - 0,96 ;AAaa -0,92 ;Aaaa - 0,76 and aaaa
- 0. The individual components of the seed storage protein patterns of
the regenerant (R) and the donor (D) were divided into two groups: one
with those occurring as expected or so, and one with those occurring significantly
otherwise. A variety of genetic factors (incompatibility systems, selection
among gametes etc.) might account for the observed difference in frequency.
Further analysis was performed on the former group (Table 1.) and the difference
between the respective frequencies of the individual components within
each pair of plants was proved significant. We did not observed new (mutant)
variants of a component, which allows us to conclude that tissue culture
conditions must be influencing only the expression of genes controlling
the synthesis of these components.
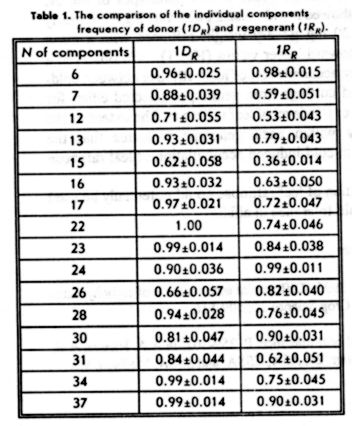
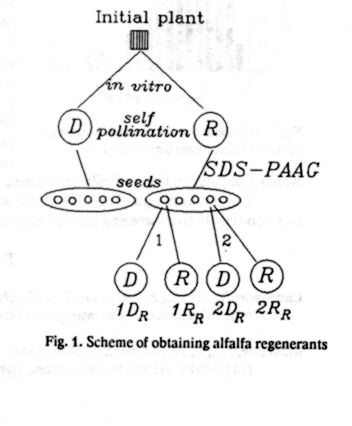
Callus Initiation and Development of Medicago
sativa L. Cultured in vivo
Guochen Yang and Marihelen Kamp-Glass
Department of Natural Resources and Environmental Design
North Carolina A&T State University
Greensboro , NC 2741 1
Plant transformation techiques have been successfully used to incorporate
particular genetic constructs to plant genome to improve disease and/or
stress resistance/tolerance for many plant species, such as alfalfa (Kuchuk
et al., 1990). In-vitro shoot regeneration protocol must be first established
and optimized in order to establish an efficient and reliable transformation
system. As a member of legume family, alfalfa is very difficult for tissue
culture, although success has been achieved on embryogenesis (Meijer &
Brown, 1987). However, limited research was done on plant growth regulators'
(PGR) effects on alfalfa callus initiation and development in order to
enhance embryogenesis and/or organogenesis (Saunders & Bingham, 1972
and 1975). Therefore, this research was initiated with following objectives:
(a) to evaluate how different PGR influence alfalfa callus initiation and
development; (b) thus to establish a foundation for further development
of shoot organogenesis protocol to assist transformation.
Seeds of cultivar WL 31 1 were used as explants. Seeds were sterilized
by soaking in 15% (0.78% NaOCl) bleach solution for 15 minutes, rinsed
3 times with sterile distilled deionized water. M.S. basal medium supplemented
with 3% sucrose and 0.7% agar, pH 5.8, plus BA at 1, 5 or 10 mg/l; zeatin
at 0.1, 1, 2, or 5 mg/l; 2,4,5-T at 0. 1, 1, 5, 10 mg/l as treatments.
Four (4) sterilized seeds were transferred to each petri dish containing
approximately 25 ml of medium of particular treatments. Petri dishes were
placed in the culture shelves with 16 hours of light provided by cool whte
fluorescent tubes at 23 + 3 °C.
BA at I or 5 mgll enhanced callus initiation and development toward shoot
organogenesis. The calli regenerated from these two treatments are compact,
solid and dark green. SNOW white calli were also noticed. BA at 5 mg/l
produced more sucll calli than at I mg/l. However, BA at 10 mg/l inhibited
callus development, only small amount of watery calli was produced.
Zeatin at 5 mg/l was the best among the concentrations tested. Large amount
of dark green, compact and solid calli was regenerated while other treatments
only produced watery and soft calli. At 5 mg/l, semi-compact, green, bright
purple, and snow white calli were also developed.
Concentration of 2, 4,5-T at 1 mgll promoted callus production, while higher
concentrations inhibited callus initiation. However, only soft calli with
sometimes snow white spots were developed from 2,4,5-T treatments. A cytokinin
is apparently needed to stimulate shoot organogenesis from such calli.
Calli regenerated can be further stimulated to induce shoot initiation,
thus to establish a regeneration protocol to assist genetic transformation.
MOLECULAR GENETICS OF A MODEL-PLANT: MEDICAGO
TRUNCATULA
T.Huguet (l), M.Gherardi (1), L.Tirichine (1),
I. Bonnin (2), G. Genier (2), J. Ronfort (2), J.M.Prosperi (2)
(1) LBMRPM, CNRS-INRA, BP27, 31326 Castanet-Tolosan Cedex, France.
(2) SGAP, INRA, Domaine de Melgueil, 34130 Mauguio, France.
Alfalfa (Medicago sativa) is a forage crop of considerable worldwide
agronomic importance having a high nitrogen fixation and protein productivity
potential. Unfortunately, both fundamental and applied research on alfalfa
is limited by its complex genetic structure (autotetraploid, allogamous
and with inbreeding depression). For these reasons, we use the mediterranean
forage legume Medicago truncatula as a model-plant to circumvent the limitations
of alfalfa and to combine genetic and molecular biological studies (Barker
et al. 1990). M.truncatula is an annual forage, diploid, autogamous plant
of low DNA content and with which transgenic plants can be routinely obtained.
M.truncatula is nodulated by well-known Rhizobium meliloti strains and
is of good agronomical value for mediterranean regions where they are utilized
to improve the soil structure, increase soil nitrogen and as a source of
winter forage. Selection of medics for introduction in mediterranean marginal
lands started in 1985 at INRA France (Prosperi 1993). A high level of polymorphism
can be observed both between and within natural populations for as well
neutral PCR markers (RAPD) as morphological traits Bonnin et al 1996 a,b).
This variability is used in breeding programs to improve characters such
as tolerance to cold, to diseases or to grazing.
In order to identify and isolate plant genes related to Rhizobium-legume
symbiosis and/or of agronomical interest, we are engaged in the construction
of a genetic map of M.truncatula. This map is based on crosses between
4 polymorphic genotypes originating from geographically different areas:
1 natural population from France, 2 from Algeria and l Australian cultivar
(Jemalong). Using three of these crosses, recombinant lines have been obtained
by single seed descent. From a single cross between Jemalong and a natural
population, we have already been able to map 50 RAPD markers, 2 isoenzymes
(PGD, PGM) and 6 known genes (rDNA, ENOD12, MTGSa, MTGSb, MTLbl, MTLec2).
This map defines 9 linkage groups (2n=16) spanning over 571 cM. Haploid
DNA content was measured by flow cytometry to be 0.58pg/lC, ensuring approximately
lMB/cM. We are presently mapping a set of symbiotic mutations (Nod++, Nod-
and Fix-) obtained by exposure to gamma rays (Sagan et al 1995). In addition,
M. truncatula is known for a long time to show a strong cultivar x Rhizobium
strain specificity. The above natural populations are now screened with
a set of wild-type Rhizobium meliloti bacterial strains.
This genetic map will be a powerful tool for the positional cloning of
symbiotic genes and for the breeding of mediterranean forage legumes as
well as for studying evolutionary processes within Medicago genus.
References
Barker, D. G., Bianchi, S., Blondon, F., Dattee, Y., Duc, G., Flament,
P.,Gallusci, P., Genier, P., Guy, P., Muel, X., Tourneur, J., Denarie,
J., and Huguet, T. 1990. Medicago truncatula, a model plant for studying
the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol.
Rep. 8, 40-49.
Prosperi, J. M., 1993. Selection of annual medics for French Mediterranean
regions. In Workshop on introducing the ley farming system to the Mediterranean
basin. 173-191. (1989) Perugia (Italy). Eds.S. Christiansen, L. Materon,
M. Falcinelli et P. Cocks.
Sagan, M., Morandi, D., Tarenghi, E., Duc, G. 1995. Selection of nodulation
and mycorrhizal mutants in the model plant Medicago truncatula Gaerth after
gamma rays mutagenesis. Plant Science,111,63-71.
Bonnin, I., Huguet, T., Gherardi, M., Prosperi, J. M., and Olivieri, I.
1996 a. High level of polymorphism and spatial structure in a selfing plant
species, Medicago truncatula (Leguminosae), using RAPD markers. American
Journal of Botany. 83(6) In press.
Bonnin, I., Prosperi, J. M., and Olivieri, I., 1996 b. Genetic markers
and quantitative genetic variation in Medicago truncatula (Leguminosae):
a comparative analysis of population structure. Genetics. In press.
Evaluation of cold tolerance in annual medics
with Potential for use in rotation with wheat on the U.S. High Plains
J. M. KRALL, R. H. DELANEY, D. A. CLAYPOOL and R. W. GROOSE
Department of Plant, Soil and Insect Sciences, University of Wyoming,
PO Box 3354, Laramie WY 82071 USA
On the U.S. Western High Plains winter wheat is most often grown in
a 2 yr rotation that involves a 14 mo fallow period. Inclusion of a self-regenerating
winter annual legume as "green fallow" could contribute to a
more economically and ecologically sustainable agroecosystem. Based on
geographical origin in cold climates, we identified 66 potentially winter-hardy
experimental lines representing 11 annual Medicago spp. These lines were
tested along with 7 commercial Australian annual medic cultivars, 2 winter-hardy
alfalfa cultivars ('Ladak 65' and 'Anik'), and 1 black medic cultivar ('George')
in trials seeded in Aug 1995 at three locations in Wyoming. M rigidula
and rigiduloides proved to be the most winter-hardy annual medic species
with 74% and 56% of lines, respectively, surviving at one or more locations
in June 1996 (Table 1). Limited survival of M. orbicularis (20% of lines)
was observed. No experimental lines or cultivars of M. aculeata, arabica,
disciformis, littoralis, polymorpha, rugosa, scutellata, tornata and truncatula
survived at any location. At Laramie, all surviving lines were flowering
on 10 June except M. orbicularis and M. sativa lines. Very low survival
(Ladak 65 only) in sandy soil at Torrington was due to desiccation in late
winter when adjacent winter wheat plants were also winter killed.

Breeding Medicago lupulina for Pasture and
Green Manure
G. V. Stepanova, E. N. Atlakova, G. P. Zatchina
William Fodder Research Institute, Lugovaja, 141740, Russia
Black medic (Medicago lupulina) - the most widespread poli morphic species
of genus Medicago, is growing in different climatic conditions. There are
spring- and winter, annual- and biannual forms. It is self-pollinating.
In Russia in the 19th century black medic was considered to be one of the
best pasturable plants. Then it have been forgotten for many years, and
only since 1989 selection work with this valuable fodder crop began in
William Fodder Research Institute. The main selected signs are: yield of
the dry matter, seeds, plant regrowth, winterhardiness, the high symbiotic
nitrogen fixation ability, resistance to the main diseases. About 100 wild
growing specimens have been used as an initial material. The methods of
breeding are: polyploidy, mutagenesis, directional selection. Black medic
samples of pasture type have been tested in grass mixture with early spring
sowing. While medic samples has been used as green manure it was inoculated
with appropriate Rhizobium strain, sowed in August and ploughed in the
following year.
The dry matter harvest of pasture mixture with different breeding medic
samples varied from 13,13 to 15,26 t/ha, with medic content from 7,83 to
13,58 t/ha for two years using. Winter hardiness of the best two samples
was 80 and 87%, others samples-4 25%. Black medic can grow in the pasture
for many years owing to self-sowing. When black medic was used as green
manure, it was cut at the end of June, then it regrown and in September
or in October it was ploughed. The dry matter harvest of testing samples
in first cutting was 3,0-3,5 t/ha. Content of nitrogen in the roots and
regrowth was 137-201 kg/ha. Breeding to increase nitrogen fixation ability
and resistance to main diseases was held in order to receive large harvest
of dry matter and seeds. Six specifical for black medic Rhizobium meliloti
strains have been created. The inoculation increased dry matter harvest
by 53,7-72,2%, seeds - 112,2-149,6%. The main disease, striking black medic
is root rot, caused by fungus of genus Fusarium. Species F.culmorum, F.
oxysporum, F. moniliformi, F.solani, F. sambicium are the most aggressive
for M.lupulina. The best breeding samples have been struck by 17,7-20,0%,
the others 26,4-45,1%.
Assignment of Medicago rigidula accessions
in the NPGS
into the two species M. rigidula and M. rigiduloides
D.C. HEFT and R.W. GROOSE
Department of Plant, Soil and Insect Sciences, University of Wyoming,
PO Box 3354, Laramie WY 82071 USA
Recently the cold-tolerant annual Medicago species M. rigidula was differentiated
into the two species M. rigidula (L.) All. and a new species, M. rigiduloides
E. Small, based on number of coils per pod and pod spine curvature where
European types (M. rigidula) have, on average, fewer coils per pod and
more hooked pod spines than Asian types (M. rigiduloides; Small et al.
1990, Can. J. Bot. 68:2607; Small 1990 ibid.:2614). Small et al. (1990)
indicate that these species are even more distinctly differentiated by
pollen morphology where M. rigidula has cylindrical pollen in contrast
to the pyramidal-triangular pollen of M. rigiduloides. Recent research
at the South Australian Research and Development Institute and the University
of Wyoming has demonstrated differences between these species in Rhizobium
specificity and winter-hardiness (Groose et al. 1996; Krall et al. 1996;
see abstracts in this volume). Groose et al. (1996) have also demonstrated
a partial barrier to interspecific fertility between M. rigidula and M.
rigiduloides. The goal of this study was to differentiate all available
accessions of "M. rigidula" in the U.S. National Plant Germplasm
System into M. rigidula and M. rigiduloides. Three plants of each line
were grown in the greenhouse. Pollen was observed at flowering and pods
traits were evaluated at maturity according to the key of Small et al.
(1990). Lines were classified as M. rigidula and M. rigiduloides on the
basis of cylindrical versus pyramidal-triangular pollen, respectively.
For lines that produced unusual pollen phenotypes or appeared to be mixed
lots, pollen of 10-20 additional plants were observed. In total, 307 ofthe
332 accessions of"M. rigidula" in the NPGS were examined. As
summarized in Table 1, the vast majority of accessions produced either
predominately cylindrical pollen (M. rigidula, 103 lines total) or predominately
pyramidal-triangular pollen ( M. rigiduloides, 192 lines total). Seven
accessions produced pollen typical of both species, as well as some pollen
grains of an intergrade phenotype. These accessions were classified as
'intermediate.' Finally, five accessions proved to be mixed lots. Four
Asian accessions included plants of both M. rigidula and M. rigiduloides
and one European accession included plants of M. rigidula and M. polynmorpha.
The intercontinental differentiation between M. rigidula and M. rigiduloides
is not as distinct as reported by Small et al. (1990). In the NPGS, 9%
of European accessions are M. rigiduloides and 15% of Asian accessions
are M. rigidula. The pod morphology of European rigriduloides accessions
was essentially identical to Asian accessions of that species. Conversely,
Asian M. rigidula accessions had pods that were nearly identical to M.
rigiduloides pods. Thus, the key to pod morphology in Small et al. (1990)
would not be useful for differentiating these species in Asia.
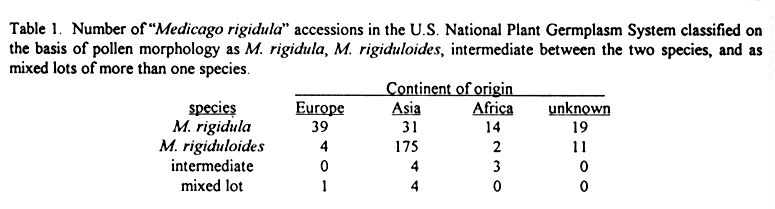
Sources of Resistance to Anthracnose in
the Annual Medicago Core Collection
N. R. O'Neill and G. R. Bauchan
Soybean and Alfalfa Research Laboratory
USDA, Agricultural Research Service
Beltsville, Maryland, 20705, U.S.A.
Recent interest in use of annual species of Medicago in the U.S. prompted
an evaluation of the diversity of germplasm that exists and in the development
of an annual Medicago core subset. This collection represents the genetic
diversity inherent in over 3200 accessions of annual Medicago (1,2). The
collection is being evaluated for sources of resistance to major indiginous
diseases, both to enhance the utilization of the species as a crop and
as a potential source of new or novel resistance genes amenable for incorporation
into adapted perennial alfalfa. Most annual medics are diploids and thus
genetically simpler to study and manipulate than alfalfa. Anthracnose caused
by Colletotrichum trifolii is a major disease in perennial alfalfa grown
in North America. Disease control is based principally on the use of resistant
varieties (3). Resistance has been associated with the production of several
pterocarpan and isoflavan phytoalexins following infection attempts in
germplasm with genes for resistance (4,5). Using standardized methods,
201 accessions from 33 species were evaluated in growth chambers for resistance
to anthracnose caused by C. trifolii. M. sativa standard check cultivars
Arc (resistant) and Saranac (susceptible) were included. Most annual medic
species were highly susceptible. Fourteen accessions representing seven
species exhibited more than 60% resistance. These were tested for inducible
defense mechanisms based on production of phytoalexins. TLC bioassays of
seedling extracts revealed the presence of fungitoxic compounds 72 h after
inoculation with C. trifolii. Medicarpin and small quantities of sativan
were produced in resistant but not susceptible or non-inoculated lines
of M. turbinata (PI 566593 from Lebanon). M. muricoleptis (PI 495401 from
Italy) produced an unidentified fungitoxic compound in inoculated seedlings.
An accession of M. truncatula produced medicarpin and sativan. Medicarpin,
sativan, vestitol, and an unidentified compound were produced in small
quantites in M. radiata (PI 459146). In all resistant species producing
phytoalexins, spores germinated and produced typical appressoria but failed
to produce primary or secondary hyphae. The inducible phytoalexin response
exhibited in accessions of M. muricoleptis and M. turbinata may account
for their resistance to anthracnose and could be exploited as a potential
source of new resistance genes. The mechanism of resistance in the remaining
twelve lines exhibiting significant anthracnose resistance appears to be
unrelated to the inducible production of phytoalexins. Because little resistance
to anthracnose was found in the core collection, resistance should be checked
and incorporated into species of annual Medicago intended for use in areas
where anthracnose is severe.
References
1. Diwan, N., Bauchan, G.R., and McIntosh, M.S. 1994. A core collection
for the United States annual Medicago germplasm collection. Crop Sci. 34:279-285.
2. Diwan, N., McIntosh, M.S., and Bauchan, G.R. 1995. Methods of developing
a core collection of annual Medicago species. Theor. Appl. Genet. 90:755-761.
3. O'Neill, N.R. 1996. Pathogenic variability and host resistance in
the Colletotrichum trifolii/Medicago sativa pathosystem. Plant Dis. 80:
450-457.
4. O'Neill, N.R., and Saunders, J.A. 1994. Compatible and incompatible
responses in alfalfa cotyledons to races 1 and 2 of Colletotrichum trifolii.
Phytopathology 84:283-287.
5. O'Neill, N.R. 1996. Defense expression in protected tissues of Medicago
sativa is enhanced during compatible interactions with Colletotrichum trifolii.
Phytopathology 86: in press.
Influence of Light on Resistance to
Vascular Wilt Diseases of Alfalfa
B. W. Pennypackerl, M. L. Risius1, J. J Volenec2, and S. M. Cunningham2
Department of Agronomy, Penn State University, and 2Department of Agronomy,
Purdue University
University Park Pennsylvania, 16802, and West Lafayette, Indiana 47907,
U. S. A.
Fungal-vascular-wilt diseases of alfalfa are managed through genetic
resistance. Resistance to Fusarium oxysporum £ sp. medicaginis is
controlled by a dominant gene and an additive gene (1). Resistance to Verticillium
albo-atrum is polygenic in U. S. commercial alfalfa but dominant in the
English variety Maris Kabul (3). Polygenic resistance to V. albo-atrum
failed under 40% ambient photosynthetic photon flux density (PPFD), indicating
carbon assimilation is critical to the expression of polygenic resistance
(2). To determine whether dominant resistance also requires carbon assimilation,
we conducted a factorial shading experiment using F. o. medicaginis to
invoke dominant resistance and V. albo-atrum to activate polygenic resistance
within clones, which avoided the problem of genetic heterogeneity between
clones. Four clones, all with dominant resistance to F. o. medicaginis,
but representing the spectrum of polygenic response to V. albo-atrum, were
used.
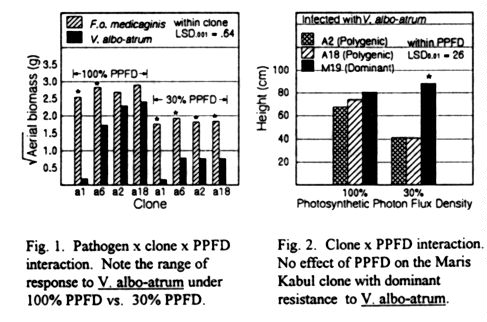
Clones were root-soaked for 15 min. in sterile water, V. albo-atrum, or
F. o. medicaginis., then grown for 6 weeks before being cut to 4-cm height
and used in the experiment; a split-plot, randomized complete block, with
3 replications. The main-plot was PPFD (ambient and 30% of ambient), and
sub plots were a 3 x 4 x 2 factorial of pathogen, clone, and sampling time.
Photosynthetic Photon Flux Density Destructive sampling occurred in weeks
3 and 5 of the S-week growing cycle. A Fig. 2. Clone x PPFD interaction.
Maris Kabul clone having dominant No effect of PPFD on the Maris resistance
to V. albo-atrum was Kabul clone with dominant included for verification
of the dominant resistance to V. albo-atrum. resistance response. The experiment
was conducted once and is being repeated. A significant pathogen x clone
x PPFD interaction was detected in aerial biomass, leaf and stem dry weight,
and height (Fig. 1). Polygenic resistance to V. albo-atrum was lost under
30% ambient PPFD, while dominant resistance to F. o. medicaginis was not
affected. A significant clone x PPFD interaction (Fig. 2) was detected
when two polygenically-resistant clones and the Maris Kabul clone with
dominant resistance to V. albo- atrum were analyzed, verifying the PPFD
insensitivity of dominant resistance. Root tissue from one polygenically-resistant
clone was analyzed for vegetative storage protein (VSP). No significant
pathogen x PPFD interactions were detected, indicating VSP was not contributing
to the PPFD sensitivity of polygenic resistance. Unlike the expression
of polygenic resistance, the expression of dominant resistance does not
appear to be contingent on carbon assimilation.
References
1. Hijano, E. H., Barnes, D. K., and Frosheiser, F. I., 1983. Crop Sci.
23:31-34.
2. Pennypacker, B. W., Knievel, D. P., Risius, M. L., and Leath, K. T.
1994. Phytopathology 84: 1350-1358.
3. Viands, D. R. 1985. Crop Sci. 25: 1096-1100.
PERSISTENCE AND LEVELS OF CROWN AND ROOT ROT DISEASES
IN ALFALFA
CULTIVARS UNDER MOWING AND ROTATIONAL GRAZING MANAGEMENT
SYSTEMS
A. Navarro and E. H. Hijano
INTA - Argentina
Alfalfa is the primary forage crop in the Pampean Region of Argentina
where approximately 5.0 million ha area grown annually. Even though alfalfa
cultivars are being tested under cutting and rotational grazing, the effects
of both management system have never been compared in simultaneous trials
using the same participants, at the same location during a 4-year period.
The objective of these experiments was to compare persistence and levels
of root diseases in alfalfa cultivars subjected to rotational grazing and
mowing.
The two trials were established in April, 1989 at Manfredi Exp. Stn, Cordoba,
Argentina. The grazing trial was a randomized complete block design having
800 m2 plots with 3 replicates. Plots were grazed with Holstein steers
using a variable stocking rate according to the available forage in a 7-day
rotational system with 35-day resting period.
The same cultivars were planted in 9 m2 plots, using a randomized complete
block design with 4 replicates and mowed at the 3rd. or 4th. day of grazing
in the "grazing" trial.
After 4 growing seasons, surviving plants were sampled and scored. In the
cutting trial, all plants from a 5 m2 cutting area were extracted. The
same area was sampled from each grazed plot, digging up plants from five
m2 subsamples randomly located within each 800 m2 plot.
Surviving plants were counted and evaluated in a 1 to 5 scale for corky
root rot and crown root rot complex severity levels.
Average surviving plants was higher under grazing (14.38 pl/m2) when compared
with the cutting (9.62 plt/m2) management system. In both experiments,
significant differences in persistence were detected among cultivars. Monarca
SP INTA and Victoria SP INTA had the highest values of surviving plants/m2
whereas Cuf 101 and WL 605 had the lowest persistence.
Average severity levels for CRRC and corky root rot were almost identical
in both trials being the ranking of cultivars, in terms of disease severity
index, quite similar.
From these results, it is concluded that the management system did not
affect differentially neither the persistence nor the ranking of alfalfa
cultivars measured as levels of crown and root diseases after 4 growing
seasons. Cultivar genotype is the main characteristic that determines persistence
or crown and root diseases levels, independently of the management system
applied for harvesting the forage.
POST-HARVEST FUNGAL RESISTANCE IN ALFALFA: CULTIVAR
RESPONSE AND
MECHANISMS
V.I Babij1, K. M. Wittenbergl, and S.R. Smith, Jr 2
Animal Science Department Plant Science Department
University of Manitoba, Winnipeg, Manitoba R3T 2N2
Alfalfa cultivars have been developed for desirable qualities such as
feed quality, winter hardiness, grazing tolerance, but no selection protocols
have been developed for post harvest fungal invasion. An effective method
for screening plants would be beneficial for development of cultivars that
have increased resistance to saprophytic organisms during field wilting
and storage. Fungal organisms, which include the genera Mucor, Absidia,
Rhizopus, Aspergillus and Humicola, develop in inadequately dried hay and
reduce hay quality. These organisms also produce spores which are a major
cause of respiratory problems for livestock and livestock producers. Preliminary
studies have suggested that certain alfalfa genotypes show resistance to
mold growth after harvest.
One objective of this study was to verify a leaf screening procedure developed
by Wittenberg et al, ( in preparation) for detection of plants resistant
or supportive of fungal growth. The screening procedure involves random
selection of alfalfa leaves from individual plants, placement of selected
leaves onto agar plates, spraying with a fungal spore suspension, incubating
at 25 degrees C, and evaluating the fungal biomass coverage on the plated
leaf material after the incubation period. Two verification studies were
conducted on individual genotypes previously identified as having low,
variable and high susceptibility to fungal growth after harvest. The first
study simultaneously compared the leaf screening procedure with mold content
of incubated stem and leaf material harvested from the entire plant. The
second study compared fungal development for these same genotypes during
field wilting and bale storage. The results of the first study indicate
that the visual evaluations from the screening procedure correspond to
the mold content of the whole plant harvested material, however minimal
fungal growth across all genotypes in the second study made comparative
evaluation impossible.
The second objective was to develop resistant and susceptible populations
using the leaf screening procedure. A population of approximately 1000
genotypes representing 22 cultivars was screened for genotypes that showed
reduced post-harvest fungal invasion for the resistant population, and
the susceptible population included genotypes that supported fungal growth.
Studies are being conducted to evaluate these populations for potential
physical, chemical and microbial characteristics associated with increased
resistance to post harvest fungal growth.
In conclusion, the association between post-harvest fungal resistance of
individual leaves in petri plates and mold growth in swaths and bales has
yet to be verified. Further research will be conducted this summer in an
attempt to relate laboratory and field resistance.
Reference
Wittenberg, K. Smith, R. and Katepa Mupondwa, F. 1995 Screening Methodology
for Post-Harvest Fungal Resistance in Alfalfa. In preparation.
Resistance in the Annual Medicago Core Collection
to Two Isolates of the Downy Mildew Fungus from Alfalfa
J. R. Yaege and D. L. Stuteville
Department of Plant Pathology, Kansas State University
Manhattan, Kansas, 66506, U.S.A.
Peronospora trifoliorum de Bary causes downy mildew of alfalfa throughout
the temperate regions of the world, but little is known about its host
range within the annual Medicago species. Using Standard Tests procedures
(2), we evaluated the reaction of approximately 50 seedlings each of 199
of the 211 accessions of the annual Medicago Core Collection (1) to P.
trifoliorum isolates I7 and I8 from alfalfa from Kansas and southern California
respectively. A wide range of symptoms and intensity of P. trifoliorum
conidium production occurred. Plants not supporting conidium production
were considered resistant and most resistant plants expressed no symptoms
or a faint stipple. However, the cotyledons of some resistant plants in
a few accessions were killed by a hypersensitive reaction to P. trifoliorum.
In 150 accessions, no plants supported conidium production. Species with
all plants resistant were M. ciliaris, M. constricta, M. coronata, M. disciformis,
M. doliata, M. granadensis, M. intertexta, M. italica, M. laciniata , M.
lesinsii , M. littoralis, M. minima, M. muricoleptis, M. orbicularis, M.
praecox, M. rigidula, M. soleirolii, M. truncatula, and M. turbinata Accessions
with susceptible plants are presented in Table 1. The number of accessions
with no susceptible plants for species listed in Table 1 were M. lupulina
5, M. murex 4, M. polymorpha 33, M. rugosa 6, and M. scutellata 13.
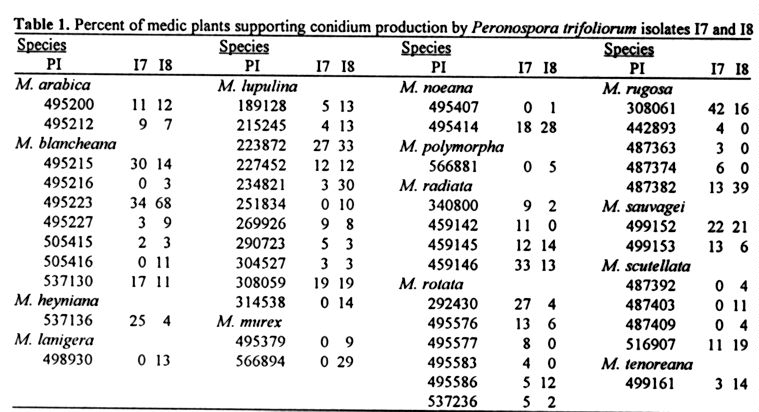
References
1. Diwan, N., Bauchan, G. R., and McIntosh, M. S. 1994. A Core Collection
for the United States Annual
Medicago Germplasm Collection. Crop Sci. 34:279-285.
2. Stuteville, D. L. 1991. Downy Mildew. D-5. In C. C. Fox, R. Berberet,
F. A. Gray, C. R. Grau, D. L. Jessen,
and M. A. Peterson (ed.) Standard Tests to Characterize Resistance in Alfalfa
Cultivars. North American
Alfalfa Improvement Conference.
Molecular markers associated with resistance
to downy mildew in tetraploid alfalfa
D. E. Obert, D. Z. Skinner, and D. L. Stuteville
Department of Agronomy, USDA-ARS, and Department of Plant Pathology
Kansas State University. Manhattan, KS 66506
The identification of molecular markers associated with disease resistance
could provide a screening method for selection of resistant genotypes,
and thus prove valuable in the development of improved cultivars. A segregating
population derived from crossing two inbred lines is not possible in alfalfa
due to its inbreeding depression (Busbice, 1968). Therefore alternative
methods of population development for genetic analysis must be employed.
Several populations differing only in one trait of interest already exist.
Two such populations, UC 123 and UC 143 differ only in that UC 143 was
selected from UC 123 on the basis of resistance to isolate I-8 of Peronospora
trifoliorum de Bary, causal agent of downy mildew in alfalfa (Lehman et
al., 1983). Since the basis of selection between these two populations
is resistance to downy mildew, any differences between the two populations,
except those occurring within limits of random allelic fluctuation, should
be associated with downy mildew resistance. The two populations, each composed
of 48 plants, were screened with 41 RAPD primers and 105 polymorphic molecular
markers were generated. Marker combinations (2-way and 3-way) were determined
and many significant marker and marker combinations were identified.
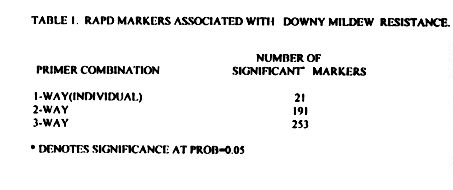
Analysis to determine genetic distances between individual plants was
performed to reduce the population size to be investigated from 96 to 72
in order to perform AFLP analysis on a fewer number of plants. AFLP analysis,
although very preliminary, appears promising since each primer combination
tested thus rar has generated over 30 molecular markers, of these markers,
an average of 12 from each primer combination were polymorphic.
Additional research is underway to confirm the association of the molecular
markers with disease resistance by the use of F, and S, progeny.
References
Busbice, T.H. 1968. Effects of inbreeding on fertility in Medicago sativa
L. Crop Sci. 8:231-234.
Lehman,W.F.,D.L. Stuteville,M.W.Nielson, and V.L.Marble. 1983. Registration
of UC 123 and UC 143 Alfalfa Germplasms. Crop Sci. 23:403.
Dynamics of Apothecial Populations of Sclerotinia
trifoliorum
P. Vincelli, J. C. Doney, Jr., and L. Wang.
Department of Plant Pathology
University of Kentucky
Lexington, KY 40546-0091.
Populations of apothecia of Sclerotinia trifoliorum were monitored during
the autumn/winter of 1991-92 (one site) and again during 1995-96 (two sites).
In all instances, apothecia first emerged between 16 Oct and 25 Oct and
were produced over a 9- to 12-wk period. Apothecial density reached a seasonal
peak within two weeks of first detection and generally was highest during
the first 4-6 wk of production, notwithstanding population fluctuations
due to prevailing weather conditions. Using weather data available for
the site in 1991-92 and for one site in 1995 96, we detected apothecia
after the mean daily soil temperature at a 10-cm depth under grass or alfalfa
dropped below 12.8 deg C followed by 0.1 cm or more of rainfall, although
stipes probably were developing prior to these conditions. Nighttime low
temperatures at or below -5°C were associated with reductions of 50-100%
in apothecial density. Studies to understand the dynamics of apothecial
populations of S. trifoliorum and the influence of weather events are continuing.
References
G., and Knight, W. E. 1982. Formation of apothecia by sclerotia of Sclerotinia
trifoliorum and infection of crimson clover in the field. Plant Dis. 1021-1023.
Valleau, W. D., Fergus, E. N., and Henson, L. 1933. Resistance of red
clovers to Sclerotinia trifoliorum Erik., and infection studies. Bull.
Kentucky Agric. Exper. Stn., Bulletin No. 341, pp 116-131.
Williams, G. H., and Western, J. H. 1965. The biology of Sclerotinia
trifoliorum Erikss. and other species of sclerotium forming fungi: Apothecium
formation from sclerotia. Ann. Appl. Biol. 56:253-260.
Disease Resistance of Australian Lucerne
Cultivars
G.C.Auricht
South Australian Research and Development Institute.
GP0 Box 397 Adelaide SA 5001, Australia
Lucerne cultivars commercially available in Australia were inoculated
with one of six different pests or diseases in separate glasshouse experiments
for detailed comparison and testing of a new design. Seed of 28 cultivars
and one check was sown in single rows in 30 x 40 cm plastic boxes. Seven
lines and one control were sown per box with three replicates giving a
total of 12 boxes per experiment. Seedlings in each row were thinned to
20 (aphid tests) or 40 (fungal diseases) then inoculated with either Blue
Green Aphid (BGA), Acrythosiphon kondoi; Spotted Alfalfa Aphid (SAA), Therioaphis
trifolii f. maculata; Anthracnose, Colletotrichum trifolii; Spring Black
Stem and Leaf Spot, Phoma medicaginis, Staganospora, Staganospora meliloti
or Phytopthora Root Rot (PRR), Phytopthera medicaginis. Experiments were
conducted and scored using techniques based on the "Standard Tests
to Characterize Alfalfa Cultivars". Trials were analysed using percentage
of resistant plants except in the case of Phoma where the mean damage score
(1 to 5) for plants in each row was analysed and BGA where both were analysed.
For SAA an analysis of variance was conducted with control rows and box
effects removed after check plants died. A square root transformation was
required before analysis of the Phoma and BGA results. BGA %Res is high
with inclusion of 3's.

Inclusion of checks increases precision, allows for box effects and
enables valid comparisons between experiments. BGA mean score gives a better
comparison than "%Res".
Alfalfa Germplasm Resources and Evaluation
of their
Disease Resistance in Semi-arid Areas of Gansu Province,
People's Republic of China
Ma Zhengyu
(Lanzhou Institute of Animal Science, Chinese Academy
of Agricultural Sciences, Lanzhou, Gansu
730050, P.R. China)
General Situation. After more than ten years elapsed, we have collected,
planted for observation, and evaluated 293 copies of various forage plant
species at fixed positions of several semi-arid areas, mountainous and
flat plain, in Yuzhong County, Central Gansu, near the Provincial Capital,
Lenzhou. Among them, there are 256 copies of legumes (occupied mainly at
a percentage of 87.37%), and the focal point of collection was alfalfa
germplasm resources, 163 copies in all (63.67% in legumes and 55.63% in
all respectively). As for germplasmic studies, with the exception of phenological
stages, agronomic characters, purity, etc., the main work of disease resistance
evaluation was done for detection of such pathological pests as, alfalfa
downy mildew, brown spot and brown leafspot diseases, and so on. Alfalfa
disease plots and germplasm propagation garden were also set up on trial.
II. Evaluation of Anti-Downy Mildew Genetic Resources in Alfalfa.
i. Method of Study. Comparative studies on manipulative inoculation in
case and meantimely direct inoculation on land test were done for pathological
description. The 5-grade statistical method was used to inspect disease
infection as usual. 2. Test Results. 68 copies of field and 36 copies of
indoor inoculation cases were tested, the results showed that the infection
rate indoor reacted fluctuately with the lowest percentage of 4.8% and
highest of 81% in accordance with their respective grades affected. The
alfalfa cultivars tested with rate of infection under 20% were as follows:
Saramas, No. 80-19 (introduced cv.), U.S. No. 2, Zhaodong (local land race),
Netherlands Lucerne, Resis, Oakland and so on; those between 20% and 50%,
Xinjiang Anti-drought, Rhizoma, Vernal, Zhaodong No. 18, Northwest A, No.
79-76 (introduced cv.), Humeng No. 18 (Local land race) and so on; as well
as those higher than 50%, only 3 in number, Tumu No. 2, French lucerne
and a local land race, Tianshui, with respective rates of 55.5-68.2%, 57.5-68.2%
and 78.0-81.0% (indoor records). Contrarily, those in showing the natural
rate of infection in field higher than 50%-81% were only two, namely, they
are Qingyang (Gansu land race) and Hetian (Xinjiang land race). 3. Final
explanation for evaluation results. Our discussion will center on disease
resistance against alfalfa downy mildew (Peronospora aestivalis), the preliminary
viewpoint is that almost and even always, the most resistant cultivars
and land races in alfalfa are those introduced from abroad, such as U.S.
No. 2, 81 85, 80-19, 81-125, Netherlands Lucerne, and so on that belonging
to the typically high-resistant and resistant model plants, but all those
local land races planted in China are susceptible. Among the 30 alfalfa
land races tested, we just have only three which are said to be highly
resistant to downy mildew, and the remained 27 are nothing but merely included
to be medium susceptible or highly susceptible. III. Sieved Type Breeding
of Alfalfa Genetic Resources.
1. Sieved Type Breeding Test in Selecting Anti-Downy Mildew Cultivars and
Land Races of Alfalfa. In the midst of 148 copies of alfalfa cultivars
and land races introduced from 14 countries abroad, we have done lots of
work on manipulative inoculation, repeated plus reversal selection and
some other methods, still up to the present, we just tentatively selected
18 out of those alfalfa cultivars and land races forementioned to be the
most productive and highest downy mildew resistant category. Acknowledgment
We heartily thank Professor Wu Renrun's Kind recommendation and disinterested
assistance to help consulate the paper to its perfection extent in the
presenting to the 35th NAAIC poster session.
Detection of Antixenosis Resistance Mechanism
to Blue Aphid
in an Argentine Alfalfa Population
J. O. Gieco, D. H. Basigalup and E. H. Hijano
EEA Manfredi, I.N.T.A., Cordoba, Argentina
Genetic variability in alfalfa regarding reaction to blue alfalfa aphid
(BAA), Acyrthosiphon kondoi Shinji, has been found; however, there are
practically no studies on identification of resistance mechanisms.
In a previous paper (1), we reported on the detection of antibiosis and
tolerance mechanisms in 36 alfalfa clones ranging from extreme susceptibility
to high resistance to BAA. Four clones were identified as primarily antibiotic,
thirteen clones were identified as primarily tolerant, and two clones simultaneously
exhibited both.
The same plant population was also tested for the presence of antixenosis,
a resistance mechanism defined as plant unattractiveness for insect food,
shelter, or oviposition. In this paper we are going to present the results
of this test.
The 36 alfalfa plants were selected after a severe BAA attack at the Manfredi
Exp. Stn.-INTA breeding nursery in 1991. All plants were cloned on vermiculite
in the greenhouse and, after 45 days, transplanted to plastic squared containers
(33 x 33 x 18 cm) filled with a 3:1 sterilized organic soil:perlite mixture
and surrounded with a transparent PVC film covered with nylon fabric. Aphid
population samples were taken from the field, purified, and increased in
the greenhouse. The test was performed under 18 24 °C, 80% RH, and
15 hr daylength conditions.
An incomplete randomized block design with 6 replications was used. Each
container received 9 different clones in a circular disposition, according
to a target-like diagram. In the center of each target, 50 adult blue aphids
were liberated; after 12, 24, and 48 hr the number of aphids on each plant
was recorded. Standardized data were used to perform a cluster analysis
(average linkage method) to group clones according to insect preference
based on pseudo F and pseudo t2 values.
Eight clusters were formed. Average aphid numbers from the most antixenotic
to the most susceptible clusters were 2.28, 2.43, 3.28, 11.50, 10.92, and
10.09 at 12, 24, and 48 hr after liberation, respectively. The most antixenotic
group included clones 2, 8, 10, 11, 12, 15, 16, 24, and 32. While clone
32 had been previously identified as antibiotic (1), in this test was also
defined as antixenotic; this is in agreement with the idea that a plant
can express more than one resistance mechanism.
REFERENCE
(1) GIECO, J. O., E. H. HIJANO and D. H. BASICALUP. 1994. Report 34th NAAIC,
Guelph, Ontario, Canada, July 10-14, p. 112.
Are Chemical Residues Sufficient in Killing or Deterring Blister Beetle
Swarms in Alfalfa?
P. G. Mulder, Jr. and K. T. Shelton
Department of Entomology, Oklahoma State University
Stillwater, OK 74078-3033
Cantharidin, the blistering agent produced by blister beetles, was first
isolated in 1810 from Lytta vesicatoria (Linn.). It is highly toxic if
taken internally. Horses are extremely sensitive to cantharidin. Approximately
lmg/kg is the minimum lethal dose for horses (Ray et al. 1980). In Oklahoma,
blister beetles complete one generation per year. Although several species
are common throughout the United States, the striped blister beetle, Epicauta
occidentalis (Werner) is the gregarious insect most commonly associated
with alfalfa in Oklahoma. Several authors have suggested means of combating
blister beetle problems (Mulder et al. 1993); however, because of the elusive
nature of the insect and unpredictability of its peak occurrence, producers
either ignore the problem or treat prophylactically just before cutting.
Unfortunately, insecticides currently labeled for control of blister beetles
in alfalfa have preharvest intervals of seven days or more. Concerned over
the immigration of swarms from adjacent areas into treated fields, producers
often choose to ignore waiting periods and harvest before preharvest intervals
have expired.
To further elucidate the subject of residual control of Sevin insecticide
on blister beetles, a greenhouse study was conducted using nearly 3,000
beetles captured in the field. Control of blister beetles, ranging from
92% to 53.4% was obtained from 1 to 120 hours after treatment with Sevin
XLR. Over the first five days, percent mortality decreased by 10.6% in
treated plants. In untreated plants, percent mortality decreased by 28%.
Higher rates of mortality in untreated plants was due, in large part, to
mortality caused by manual infestation. Based on analysis of forage consumed
during the trial, data suggest that beetles are not only effectively killed
by chemical residues but are also deterred from consumption of foliage.
Information from this study and two extension publications was presented
at approximately six regional meetings across Oklahoma in 1995. Over 500
producers had an opportunity to ask questions concerning their alfalfa
harvest management practices.
References
- Mulder, P. G., Shawley, R. V., and Caddel, J. L. 1993. Blister beetles
and alfalfa. Oklahoma State University, Oklahoma Cooperative Extension
Service Fact Sheet No. F-2072.
- Ray, A. C., Post, L. O., Hurst, J. M., Edwards, W. C. and Reagor,
J. C. 1980. Evaluation of an analytical method for the diagnosis of cantharidin
toxicosis due to ingestion of blister beetles (Epicauta lemniscata) by
horses and sheep. Am. J. Vet. Res. 41: 932-933.













