ALFALFA BIOTECHNOLOGY SYMPOSIUM
Alfalfa Chromosome Mapping: a Progress Update
and Discussion on Utility
J. H. Bouton .......................................................................63
Analyzing Large Sets of Molecular Marker Data - D. Z. Skinner ...........................64
Development and Utilization of an Agrobacterium-Mediated
Transformation System
for Alfalfa - Deborah A. Samac and Carroll P.
Vance.................................65
Genetic Manipulation of Disease Tolerance
and Lignin Quality - Richard A. Dixon,
Sameer A. Masoud, Ken Korth and Vincent J.
H. Sewalt.................................................66
Potential for Phytoalexin Engineering in Alfalfa - N. L. Paiva..........67
Alfalfa Chromosome Mapping: a Progress Update and Discussion on Utility
J. H. Bouton, Department of Crop and Soil Sciences, University of Georgia, Athens, GA 30602
In diploid alfalfa, genetic maps have been developed based on molecular
markers (1,2,4). These maps have potential for utilization in alfalfa improvement
programs for the following practical approaches: marker assisted selection,
cultivar identification, selection of parents for synthetics, and comparative
mapping.
In marker assisted selection, genes of agronomic importance may be "tagged"
to molecular markers closely linked to them. Selection is then based on
the marker(s). This is useful during backcrossing and to facilitate introgression
of desired genes from wild or less agronomically acceptable species and
can even be used for traits controlled by several genes (e.g. quantitative
trait loci or QTL). A good example of this approach is the report of tagging
aluminum tolerance genes from the coerulea subspecies and their possible
use to introgress this important trait into cultivated alfalfa (5). In
cultivar identification, one is actually trying to "fingerprint"
a cultivar for patenting rights. This approach appears to have limited
use due to the amount of heterogeneity within each cultivar where plants
from different cultivars can show a closer relationship than plants from
the same cultivar. The finding that using molecular markers to maximize
genetic dissimilarity among tetraploid plants correlated with higher yield
among their single crosses increases their potential use for selecting
the best parents for synthetics (3). Finally, the high degree of hybridization
between white clover (Trifolium repens L.) DNA and alfalfa probes from
the Univ. of Georgia alfalfa map allowed construction of a white clover
genetic map which demonstrated a great deal of evolutionary rearrangement
for chromosomes of the two species (S.M. Wright, 1996, unpublished data).
This type of comparative mapping may have utility in identifying homologous
QTLs for traits such as yield which could then be maximized in alfalfa.
The main problems associated with using molecular markers in alfalfa are:
(a) Virtually all applications of the existing alfalfa maps are based on
the ability of transferring the diploid maps to segregating tetraploid
populations. The use of microsattelites may have utility for map construction
at the tetraploid level thereby bypassing this problem (N. Diwan, 1996,
unpublished data). (b) Alfalfa's extreme heterogeneity increases the chances
of new alleles being present at tile locus of interest. Thus, the utility
of marker assisted selection may be reduced when multiple alleles exist
or new alleles are brought into a population of interest.
References
Brummer, E. C., Bouton, J. H., and Kochert, G. 1993. Development of
an RFLP map in diploid alfalfa. Theoretical and Applied Genetics. 86: 329-32.
Echt, C. S., Kidwell, K. K., Knapp, S. J., Osborn, T. C., and McCoy, T.
J. 1994. Linkage mapping in diploid alfalfa (Medicago sativa). Genome 37:61-71.
Kidwell, K. K., Bingham, E. T., Woodfield, D. R., and Osborn, T. C. 1994.
Relationships among genetic distance, forage yield and heterozygosity in
isogenic diploid and tetraploid alfalfa populations. Theoretical and Applied
Genetics. 89:323-328.
Kiss, G.B., Csanadi, G., Kalman, K., Kalo, P., and Okresz, L. (1993) Construction
of a basic genetic map for alfalfa using RFLP, RAPD, isozyme and morphological
markers. Molecular and General Genetics. 238: 129-37.
5 Sledge, M K., Bouton, J H., Tamulonis, J., Kochert, G., and Parrott,
W.A. 1996. Aluminum tolerance
QTL in diploid alfalfa. Report 35th North American Alfalfa Improvement
Conference. (In press).
Analyzing Large Sets of Molecular Marker
Data
D. Z. Skinner
USDA-ARS and Agronomy Department,
Kansas State University, Manhattan, Kansas, 66506-5501, U.S.A.
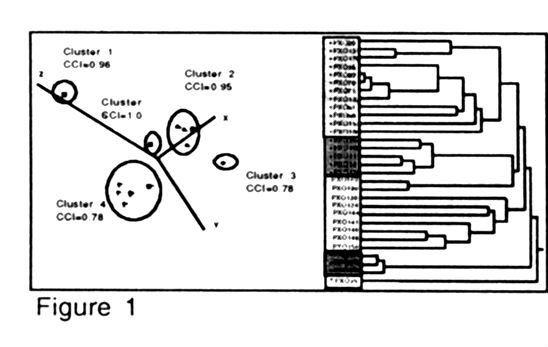
Modern techniques allow the generation of hundreds of genetic markers from single individuals isolated from populations of biological organisms. Presented here is a method of analyzing the resulting large data sets to discover the number of genetic lineages represented. The steps in the analysis are to carry out a: (1 ) Relative Apparent Synapomorphy (RAS) analysis to determine if the data are nonrandom [Lyons-Weiler et. a/., 1996]; (2) calculation of genetic distances between isolates; (3) multiple correspondence analysis (MCA). of the isolates based on the distance data; (4) cluster analysis and clustering statistics to find distinct groups of isolates (potential lineages); (5) calculation of average genetic distances within and between clusters to determine if the clusters found contain substantially less variation than the entire data set; and (6) statistical test of cluster robustness using resampling methods, resulting in the generation of a cluster consistency index (CCI). Observation of the assigned clusters in 3-dimensional space (plot of the first three dimensions from the MCA), resampling analysis to determine the reproducibility of the clusters, and average distances between and within the clusters are used to determine whether the clusters found are likely to be meaningful. The cluster method used depends on the data set under investigation and must be determined empirically. In the analysis of ideal data sets, all isolates are assigned to a lineage in one cluster analysis step. However, complex data sets may require several sequential analysis steps, i.e., as one robust cluster is discovered, it is removed from the data set and the six-step analysis is carried out on the remaining isolates. Different cluster analysis methods may be needed to discover additional clusters. The RAS analysis is used at each step to determine whether meaningful, nonrandom variation is present in the remaining isolates.

Visual display of the lineages is achieved with three-dimensional graphs
from the MCA analysis, or dendrograms with the lineages shaded (Fig. 1).
Often, the six step analysis defines lineages that are not obvious from
the dendrogram; the CCI indicates the reliability of the lineage assignments.
SAS programming to carry out the analysis is available from the author.
Reference Lyons-Weiler, J., G.A. Hoelzer, and R.J. Tausch. 1996. Relative
Apparent Synapomorphy Analysis (RASA) I: the statistical measurement of
phylogenetic signal. Molecular Biology and Evolution (in press).
Development and Utilization of an Agrobacterium-Mediated
Transformation
System for Alfalfa
Deborah A. Samac1 and Carroll P. Vance2
USDA-ARS-Plant Science Research, Department of Plant Pathology, 2Department
of Agronomy and Plant Genetics
University of Minnesota, St. Paul, MN 55108
A highly efficient transformation system based on co-cultivation of
leaf piece explants with Agrobacterium tumefaciens was developed for a
clone selected from the alfalfa variety Regen SY. Plants were first selected
for rapid regeneration in tissue culture and susceptibility to transformation
by A. tumefaciens. Three different commonly used disarmed strains of A.
tumefaciens were tested: LBA4404 (octapine type), AGL-1 ("supervirulent"
agropine type), and ABI (nopaline type). Each strain contained a plasmid
with chimeric genes for a selectable marker, neomycin phosphotransferase
(NPTII), and a scorable marker, Beta-glucuronidase (GUS). The efficiency
of transformation was highly dependent on the strain used. No GUS positive
embryos were produced with strain AsI, a moderate number were obtained
using AGL-1, while high numbers of transformed embryos were produced using
LsA44o4. Extensive explant necrosis was often observed when using strains
AGL-1 and ABI. The optimal length of co-cultivation using strain LBA4404
was seven days. Plants are transferred to soil approximately 9-12 weeks
after cocultivation.
Susceptibility of plants to transformation by wildtype oncogenic strains
of A. tumefaciens was highly correlated with dormancy group. Stems of two-week
old plants from the nine diversity groups were inoculated with strains
A208 (nopaline type), A348 (octapine type), A281 ("supervirulent"
agropine type) and A136 (avirulent). For each of the virulent strains there
was a positive correlation (P=0.05) of resistance to tumorigenesis with
dormancy and all strains behaved similarly. Plants from M. falcata and
Ladak were most resistant with 92-90% of plants forming no tumors averaged
over the three strains. Plants from Chilean and African populations were
most susceptible with an average of 68% of plants forming tumors. Tissue
explants of two-week old seedlings from the nine diversity groups were
tested for transformation by the three disarmed strains. The strong correlation
between dormancy and resistance to oncogenic strains was not observed with
disarmed strains. However, for most germplasms, there was a strong germplasm-strain
interaction. More individual plants were transformed by strain LBA4404
than with AGL-1 or AsI.
The alfalfa transformation system has been used to analyze the expression
patterns of alfalfa promoters involved in ammonia assimilation in nodules
(asparate aminotransferase, AAT; NADH-glutamate synthase, NADH-GOGAT; phosphoenolpyruvate
carboxylase, PEPC; and asparagine synthase, AS) and to identify elements
required for expression in nodules. Expression patterns of GUS controlled
by these promoters are highly correlated with in situ RNA expression for
the corresponding genes. Transgenic alfalfa plants expressing antisense
RNA of AAT, NADH-GOGAT, PEPC and AS have been produced. Expression of antisense
NADH-GOGAT using the AAT2 promoter reduces enzyme levels approximately
50% of normal levels, causes male sterility, and decreases cold tolerance
of plants. The transformation system is currently being used to engineer
alfalfa for bioremediation and for increased leaf retention.
Genetic Manipulation of Disease Tolerance And Lignin Quality
Richard A. Dixon, Sameer A. Masoud, ICen Korth and Vincent J.H. Sewalt.
Plant Biology Division, Samuel Roberts Noble Foundation, P.O. Box 2180,
Ardmore, Oklahoma 73402.
We have produced genetically transformed alfalfa lines in which a glucanase
gene normally expressed in roots in uninfected plants, and in leaves in
response to fungal infection, is now constitutively expressed at high levels
throughout the plant. These plants show reduced disease severity following
infection by the oomycete root pathogen Phytophthora megaserma f. sp. medicaginis.Transgenic
alfalfa lines constitutively expressing a rice chitinase gene, or a combination
of rice chitinase and alfalfa glucanase, did not show reduced susceptibility
to a range of pathogenic fungi, including P. medicaginis. Field testing
of these transgenic lines is in progress. Previous studies in our laboratory
have led to the cloning of a number of alfalfa genes encoding enzymes of
lignin biosynthesis, specifically phenylalanine ammonia-lyase, cinnamate
4-hydroxylase, caffeic acid 3-O methyltransferase, and caffeoyl CoA 3-O-methyltransferase.
Reducing the level of lignin in forage crops such as alfalfa should lead
to an increase in forage digestibility. We have investigated a range of
molecular strategies for reducing lignin content, and altering lignin monomer
composition, in a tobacco model system. Down-regulation of L-phenylalanine
ammonia-lyase (PAL) by epigenetic sense suppression results in decreased
lignin levels but a significant increase in syringyl/guaiacyl ratio, whereas,
surprisingly, down regulation of cinnamate 4-hydroxylase (C4H) using an
alfalfa C4H antisense transgene leads to reduced lignin with decreased
S/G ratio. Expression of an alfalfa caffeic acid 3-O-methyltransferase
antisense transgene leads to a reduction in lignin content and increased
S/G ratio. There is a negative curvilinear relationship between lignin
content and in vitro digestibility of neutral detergent fiber from stems
of a series of isogenic tobacco lines differing in lignin content through
genetic manipulation. The above studies form a basis for targetted genetic
modification of lignin in forage species. Using antisense constructs with
sequences derived from alfalfa caffeoyl CoA 3-O-methyltransferase, caffeic
acid 3-O-methyltransferase, or both sequences in the same construct, we
are attempting to determine whether the apparently parallel pathways for
monolignol formation at the level of the free acids or their CoA thioesters
are functionally redundant. We will present preliminary data for the effects
of expression of COMT and CCOMT antisense transcripts on lignin quantity,
lignin quality, and digestibility in transgenic alfalfa.
Potential for Phytoalexin Engineering in Alfalfa
N.L.Paiva
Plant Biology Division, The Samuel Roberts Noble Foundation
P.O. Box 2180, Ardmore, OK 73402 USA
Alfalfa (Medicago sativa) produces the pterocarpan phytoalexin (-) medicarpin
in response to attack by many fungal pathogens. While the growth of some
alfalfa pathogens is inhibited by medicarpin, many pathogens appear to
have evolved to be insensitive to medicarpin or to protect themselves by
degrading medicarpin. Several literature reports demonstrate that pathogens
are often more susceptible to phytoalexin structures which are not produced
by their natural host(s). Thus, introducing new phytoalexin structures
into alfalfa could generate plants which would be more resistant to pathogens.
We are identifying suitable "target" phytoalexins, isolating
structural genes from other species for introduction into alfalfa, and
identifying promoters which would be useful in driving the expression of
these genes in alfalfa. Using agar plate bioassays, we have demonstrated
that even small changes in the structure of medicarpin can greatly increase
its inhibition of fungal mycelial growth (1). For example,(+)-medicarpin
is more toxic than (-)-medicarpin to several important pathogens. Alfalfa
accumulates only (-)-medicarpin, via a pathway involving (R)-vestitone
reductase, while other legumes accumulate (+)-medicarpin, using an (S)-vestitone
reductase. We had first discovered (R)-vestitone reductase in alfalfa cell
cultures, and have since gone on to clone the (R)-vestitone reductase gene
from alfalfa (2). We are now using probes generated from the work in alfalfa
to clone an (S)-vestitone reductase. Other examples of more potent phytoalexins
would include the medicarpin derivatives (-)-homopisatin (1) and prenyl-medicarpin
(3). Homopisatin was much more toxic than (-) medicarpin to 5 of 9 pathogens
tested, and could be synthesized from medicarpin by the action of a 6a-hydroxylase
and a methyltransferase (1). Preliminary bioassays have indicated that
C10-prenylated medicarpin is very inhibitory at concentrations where (-)-medicarpin
is almost inactive; we are trying to clone a suitable pterocarpan prenyltransferase
from green bean (Phaseolus vulgaris) (3). To drive these and other phytoalexin
modifying genes, we have isolated the alfalfa isoflavone reductase gene
promoter. This promoter drives strong expression in only those cells which
are actively synthesizing medicarpin, such as those surrounding pathogen
infection sites (4); we have identified the elements which control the
pathogen activation of this promoter, and we are comparing this and other
promoters in metabolite-engineering studies.
1. Blount, J.W., and Paiva, N. L., 1996. The natural alfalfa phytoalexin
(-)-medicarpin and structurally-related compounds as inhibitors of phytopathogenic
fungi; Physiological and Molecular Plant Pathology, submitted.
2. Guo, L., and Paiva, N.L., 1995. Molecular cloning of Alfalfa (Medicago
sativa L.) vestitone reductase, the penultimate enzyme in medicarpin biosynthesis.
Archives of Biochemistry and Biophysics, 320:353-360.
3.Guo, Z., and Paiva,.N.L., 1996. Toward metabolic Engineering of alfalfa
for production of prenylated isoflavonoid/pterocarpan phytoalexins. Poster
presented at the 35th NAAIC meeting, Oklahoma City.
4. Miao, B., and Paiva,N.L., 1996. Cis-elements and Trans-acting factors
required for the regulation of alfalfa isoflavone reductase. Poster presented
at the 35th NAAIC meeting, Oklahoma City.