Breeding for Heterosis Using Traditional and Marker-Assisted Methods
E. Charles Brummer, Diane Luth, and Heathcliffe Riday
Iowa State University
brummer@iastate.edu
Abstract
Heterosis–the improvement of progeny over parents for yield or other traits–results when particular genotypes are crossed. Heterosis has been documented in alfalfa on numerous occasions, and recently, I proposed a breeding scheme to capture partial heterotic gains through the production of semihybrid cultivars. Keys to the success of this method are the identification and maintenance of heterotic groups and the subsequent improvement of populations both per se and for the interpopulaiton cross. Field experiments need to be conducted to identify heterotic groups, although identifying molecular markers linked to key "yield genes" could help differentiate populations. We have developed an F1 tetraploid intersubspecific population to begin to identify genomic regions important for yield. Four replications of five clones of each genotype were planted at two Iowa locations in 1998. First harvest yield showed substantial heterosis in 1999, with the F1 mean approximately 150-175% higher than the higher yielding parent, and individual genotypes within the F1 population yielded as much as 300% of the parents. A number of QTL for first and second harvest yield were identified through genetic mapping. The QTL of largest effects were located on linkage group 4b, the same group that showed extreme excess heterozygosity in a diploid F2 population. Alleles improving yield were contributed by both ABI408 and by Wisfal-6, suggesting contrasting genetic palettes in the two subspecies and further supporting the suggestion that the two subspecies of alfalfa represent heterotic groups and should be independently selected for agronomic traits. Finally, manipulation of fall dormancy and winterhardiness will be necessary to make full use of various germplasms in semihybrid cultivar development, and toward that goal, we found little evidence for a correlation between the two traits in our F1 population. Our results suggest the traits can be independently selected, but quickly manipulating them may need transgenic approaches.
Introduction
Improving the yield of alfalfa represents a major goal of breeding programs, yet only limited improvement is evident from either university yield trial data or from historical agricultural statistics. Three general methods can be suggested to improve the genetic yield potential of alfalfa: (1) direct selection for yield, through recurrent phenotypic selection using some form of progeny testing, (2) methods to capture partial heterosis, either true or semihybrids (Brummer, 1999), and (3) for northern areas, improving fall growth. Combining all three methods would be ideal, and I argue that all three are possibly attainable.
Capturing heterosis is perhaps the most important of the three, since it has so far met with little success. In order to realize heterotic yield gains, heterotic groups–genotypes or populations that produce high yielding progeny after crossing–need to be identified. Once they are identified, breeding methodologies need to be available to simply and easily integrate heterotic groups with current seed production capabilities, and care must be taken to avoid mixing the groups once their intercross progeny’s high yield has been recognized. However, selection within each heterotic group must be practiced in such a way to maintain or improve pest resistances, winterhardiness, forage quality, etc. without eroding the beneficial complementarities between populations. Interpopulation improvement programs–such as reciprocal recurrent selection–are not as amenable to routine use in alfalfa as in corn; much more consideration of potential breeding schemes is necessary to fully capitalize on heterotic groups that have been identified. Manipulation of fall growth and winterhardiness in a facile manner would expedite the development of heterotic groups, as non-adapted germplasm could be engineered for persistence. Throughout all these considerations, molecular markers can play roles to aid in selecting desirable plants (both for yield and for winter survival) to enhance heterotic groups and to enhance inter-population hybrids.
Heterosis refers to the advantage that progeny exhibit over their parents. Only certain crosses express heterosis, which is caused by a combination of partial to complete dominance at loci controlling the trait and of different desirable alleles or allele frequencies in the populations being crossed. Though heterosis can be measured for any trait, I confine discussion in this paper to forage yield.
Heterotic patterns
Four or five heterotic groups may be suggested within North America: Canadian and northern US very dormant germplasm (usually including subsp. falcata), semi-dormant germplasm of the upper Midwest and of the Northeastern US, and non-dormant populations in the desert Southwest. Intermediate populations from Kansas and Oklahoma may represent further groups or may be an amalgam of semi- and non-dormant groups. Another possible source of heterotic groups may be germplasm developed in-house by different commercial companies over the past 30-40 years, but preliminary evidence suggests these populations may be rather similar, at least in the Midwest (Riday and Brummer, this conference).
Other potential heterotic groups may exist among germplasm, either wild or improved, from eastern Europe, Italy, Spain, France, and North Africa. Much more research needs to be conducted to empirically determine if populations from these disparate regions will produce heterosis with each other or with US germplasm. We are in a critical period for heterotic group definition and maintenance: as cultivar development becomes more and more biotechnology driven, the ability to maintain separate genetic populations will become harder as introgression of transgenes into various populatiosn accelerates. Mixing germplasm into "melting pots" will destroy our ability to easily extract heterotic combinations of genotypes for future cultivar development. Although heterotic groups could be developed through careful breeding, starting with well defined groups is both faster and easier.
Breeding schemes to capture heterosis
Breeding semihybrid cultivars offers one potential means of capturing some heterosis (Brummer, 1999). Success of this scheme relies on the use of heterotic groups. Basically, semi-hybrids can be developed by planting selected plants from two (or more) heterotic groups in the same seed field. If only two populations are included, roughly half of the seed will be of (inter-population) hybrid origin; adding more populations will increase the amount of hybrid seed, and decrease the intra-population contribution. If heterotic groups are identified, they should be improved independently and intercrossed only for testing or for certified seed production so that new semi-hybrids can be repeatedly created.
This scheme is conceptually similar to that proposed by others (Kehr et al., 1961; Rotili et al., 1996 and this conference), who suggested that the parents on each side be inbred. I do not deny the usefulness of inbreeding to purge the genetic load (see Groose et al., this conference), but using inbred parents will result in undesirable inbreeding depression in the non-hybrid seed produced during semihybrid production. Further, developing deeply inbred parents is unlikely at the present time, so heterozygous parents are most likely to be used, which may not result in progressive heterosis (Dudley, 1964). Finally, other proposals neglect to identify heterotic groups, which will lessen the realized heterosis of the resulting semihybrid.
Phenotypic recurrent selection (PRS) has been widely used by alfalfa breeders throughout the 20th century, and its use has resulted in populations that are highly resistant to many diseases, insects, and nematodes, have acceptable yield and forage quality, and carry a decreased genetic load. The widespread use of PRS has in effect formed a few large breeding populations, for example, midwestern U.S. semi-dormant germplasm that provide little opportunity for increasing yields through hybrid development.
The implicit assumptions underlying the success of the semihybrid breeding method are that improvements both of heterotic populations per se and of the intergroup crosses can be effected. Recurrent selection to improve various agronomic characteristics should be initiated in populations that express heterosis with current commercial germplasm. One of the major alternative heterotic groups to combine with the midwestern semidormant germplasm is subsp. falcata (Brummer, 1999). However, falcata suffers from a number of deficiencies, including slow regrowth, early fall dormancy, and multiple disease susceptibilities. It also has yellow flowers which will impact pollination and seed production practices. Beginning a recurrent selection program in several different falcata populations will provide germplasm that will be more acceptable for combining with elite material. Though such an approach is necessarily long term, without enhancement efforts, wild germplasm will likely never be used. Of course, concurrent continued selection within elite populations should be practiced, particularly since semihybrid cultivar development is probably not likely in the near future.
Second, simple methods to improve interpopulation heterosis are needed. Reciprocal recurrent selection has been used successfully in maize breeding, but the difficulty of making and evaluating many crosses in alfalfa makes this method unattractive. One possibility is to produce topcross seed of each population by planting a number of individuals from one population in semi-isolation from each other along the edge of a large seeding of the other population. The resulting topcross families could be evaluated in single row plots at two locations to select the best parents to intercross for the next cycle. Once again, selecting the best individuals within the topcross family would not be desirable, as this would result in mixing heterotic groups. The topcross system as outlined could be practical for commercial breeding programs, and empirical data suggest that topcross evaluation would improve interpopulation performance (Rowe and Hill, 1981). A further alternative strategy could be to select within each population for two cycles and go to interpopulation selection only every third cycle; though this might limit interpopulation gain, it could be much faster overall. Additionally, the development of semihybrids also results in the production of intrapopulation crosses, so intrapopulation selection would help raise the performance of this portion of the semihybrid.
Molecular marker-assisted selection
Molecular markers could be integrated into the system by aiding in selection for agronomic traits within each heterotic group, by improving selection of parents for the interpopulation cross, and by improving the heterotic populations for cross performance. If marker loci at or closely linked to genes for yield could be identified, then divergent selection at those loci for contrasting alleles in each heterotic population may help improve the interpopulation hybrid without the necessity of rigorous interpopulation selection. This idea sounds fantastic in light of the limited molecular marker research currently available for alfalfa (Brummer et al., 2000).
To begin to test the utility of markers in alfalfa improvement, we are mapping a tetraploid alfalfa F1 population for yield. Mapping F1 populations results in maps for each parent because recombination and segregation is observed within each heterozygous parent (Ritter et al., 1990). In our case, this results in subspecies specific maps that should be interesting genetically and evolutionarily. In order to map tetrasomic tetraploids, we score individual alleles contributed by either parent that segregate in the progeny as present or absent. An allele present in only one copy in only one parent will segregate 1:1 for presence:absence in the F1 progeny; these alleles are referred to as single dose restriction fragments, SDRF (Wu et al., 1992) or more generically single dose alleles, SDA (Diwan et al., 1999). By analogy, alleles present in two copies of only one parent are called double dose restriction fragments (DDRF) or alleles (DDA), which segregate 5:1 in the F1 population. Linkage analysis results in the development of homologue specific linkage groups (Grivet et al., 1996); in our case, we will develop 32 maps for each parent (4 homologues x 8 basic chromosomes). The homologue specific maps can be linked together using multiple SDRF alleles from a single marker (probe or primer) or DDRF to produce a consensus or composite linkage group for each chromosome (Grivet et al., 1996).
Mapping agronomic traits, then, depends on loci controlling the trait being heterozygous in one or both parents. If a quantitative trait locus (QTL) is closely associated with a marker locus, then the QTL can be detected using single factor analysis of variance or other means if the QTL allele that is associated with a SDRF or DDRF has a large effect. What is being analyzed are F1 individuals that either carry the SDRF (and associated QTL allele) or do not carry that allele from Parent A. Within each class of progeny, the other three alleles from Parent A are present in equal frequencies. Similarly, both progeny with and without the SDRF carry two of the four alleles contributed by the other parent. Thus, a QTL will be detected if an allele associated with the marker has a strong effect in combination with all other alleles of that parent, and with all alleles of the other parent.
Mapping yield in a tetraploid F1 population
The F1 population consists of 200 individuals derived from genotypes of the two subspecies of cultivated alfalfa: a semi-improved plant of M. sativa ssp. falcata (FD = 2) from the WISFAL germplasm (Bingham, 1993) and an elite genotype of M. sativa ssp. sativa, ABI408 (FD = 3), a gift from ABI Alfalfa; both parents are highly heterozygous. This cross is segregating for numerous traits, including flower color, stem and leaf morphology, flowering time, forage yield, selfed seed production, and numerous disease resistances. The parents and population was clonally replicated and planted into field trials at two Iowa locations (Ames and Nashua) in 1998. Two additional locations, Manhattan, KS (in collaboration with Paul St. Amand) and Ithaca, NY (in collaboration with Don Viands) were planted in 1999. Five clones of each genotype were planted into plots within a 14 x 15 quadruple a-lattice at each location. Numerous traits are being evaluated in this population, but only forage yield will be discussed here.
The preliminary yield results for 1999 indicate that the F1 population produced significant heterosis at the first harvest (Table 1). Ames had higher yields than Nashua, although the population performance relative to the parents was greater at Nashua. The mean performance of the population was 150-175% more than the parents, which yielded similarly. However, some genotypes in the F1 population yielded over 300% of the parents. Very low yielding genotypes were also present. Thus, this sativa x falcata cross exhibits significant heterosis and transgressive segregation in both directions. Second harvest yields were considerably reduced from first harvest, and the population mean was not different from the higher yielding parent, ABI408. Some individual genotypes outyielded the ABI408 parent by as much as 200%.
Our linkage maps are being developed with restriction fragment length polymorphism (RFLP) markers used to develop other alfalfa maps (Brummer et al., 1993; Echt et al., 1994; Tavoletti et al., 1996; Brouwer and Osborn, 1999). Because of the difficulty developing tetraploid maps, we have so far only produced small linkage groups of 2-4 markers, with many markers unlinked. Preliminary evidence suggests that the linkage order in both tetraploid parents is similar to that seen in the diploid maps, but comprehensive statements on the order between parents, or the congruence of our map with others is not yet possible. However, extensive maps are not necessary to test single markers for association with QTL using ANOVA.
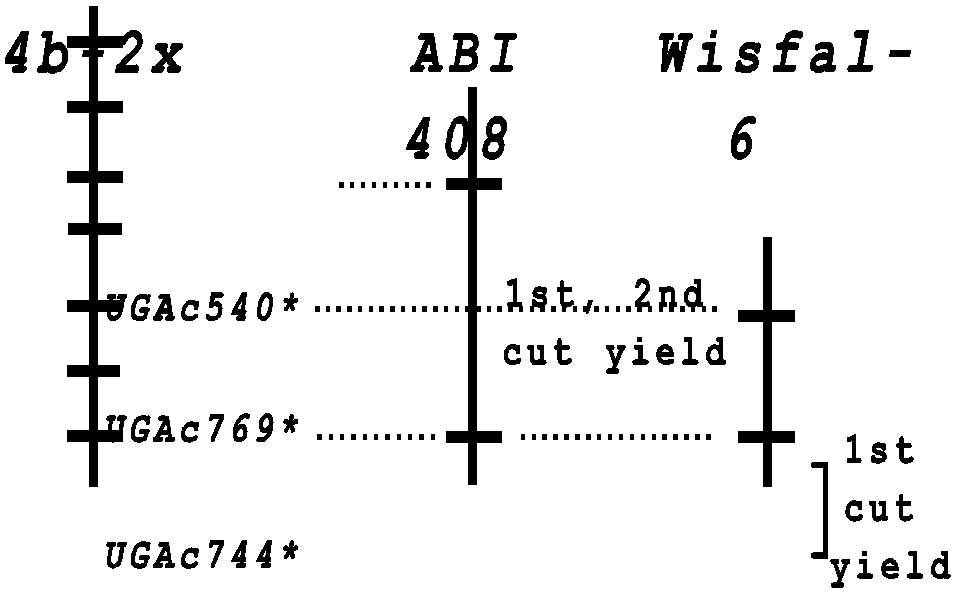
Linkage group 4b on the Univ. of GA genetic map (Brummer et al., 1993; Brummer et al., 2000) contained highly distorted segregation in a diploid F2 population, with more heterozygous individuals than expected. Excess heterozygotes suggested that this chromosomal region was involved with inbreeding depression; exposure of homozygotes upon selfing was lethal. In both parents, we detected markers having significant (P < 0.01) associations with both first and second cut yield that mapped to this region (Fig. 1). Though the data are preliminary, they offer compelling evidence that linkage group 4b contains loci important to fitness, and hence, yield. We detected a number of other QTL, though more data are needed before conclusive statements about their effects are made. However, both parents contributed QTL alleles for enhanced and for reduced yield, again suggesting that these parents have contrasting genetic palettes, having alleles that complement each other to produce higher yielding progeny.
As we get more complete results from this study, we can begin to use the marker information to select parents for synthetic cultivars and for complementarity among populations to semihybrid cultivars. Assessments of allelic diversity among genotypes may be more usefully related to yield if the markers evaluated are linked to QTL controlling yield (Charcosset et al., 1991; Bernardo, 1992). The validity of this assessment in highly heterozygous alfalfa populations with many alleles is unclear, but considering only loci with some relevance for yield is almost surely an improvement to random genome scans. Ultimately, of course, we would like to identify the genes responsible for yield to understand the genetic basis of heterosis. Of particular interest is the genetic architecture of yield QTL, i.e. the relationship between alleles, loci, and linkage blocks. The relative importance of additivity, dominance, and overdominance at particular loci (and among various alleles) as well as the role that interlocus epistasis plays in yield needs to be assessed.
Manipulating winterhardiness and dormancy
Many populations representing potentially new germplasm sources for a particular geographic region lack either the winterhardiness or the photoperiod insensitivity to be integrated with currently cultivated breeding populations. Thus, one of the bigger needs in order to make a variety of germplasm sources available for hybridization is some means to manipulate their winterhardiness and dormancy responses. The most straightforward means of doing this is through traditional breeding. At Iowa State, we are currently conducting selection both for less dormancy in subsp. falcata and for more winterhardiness in non-dormant subsp. sativa (Brummer, 1999). Certainly some improvement can be made in both populations, though whether they can reach commercially viable levels is unclear.
Many studies have suggested that fall dormancy and winterhardiness are inextricably linked (e.g. Schwab et al., 1996), but other studies show no clear relationship between the traits (Daday, 1964; Busbice and Wilsie, 1965). In our F1 population discussed above, we have examined the relationship between plant height measured in the fall and winter injury, two common measures of fall dormancy and winterhardiness, respectively. The populations were evaluated at both Ames and Nashua, IA for fall plant height in October 1998 and for winter injury in April 1999 (Brummer et al., 1999). In this populations, fall height and winter injury were not associated (Table 2). The genetic correlation between the traits was low but negative, suggesting that taller plants in the fall have less winter injury. Ours is the first study to evaluate (i) replicated plots of individual genotypes within a segregating population and (ii) a population in more than one environment.
These results suggest that breeding for both increased fall growth and decreased winter injury will be possible within this population, supporting others who have made similar suggestions (Daday, 1964; Busbice and Wilsie, 1965; McCaslin et al., 1990; McCaslin and Helming, 1995). Selection will likely not be simple, particularly on a single plant basis, as suggested by the low heritabilities we observed on a plot basis (Table 2), especially since our plots consisted of five clones. This population has a limited range of variation however; none of the F1 genotypes has fall growth similar to a non-dormant plant. In order to truly manipulate the germplasm, we may need a sequential breeding process, where increased winterhardiness or decreased fall dormancy are integrated in steps into new populations. This suggests that manipulating these traits will take a considerable time and may not permit easy integration of various germplasm sources into semihybrid production.
A second approach is through germplasm engineering. Many genes related to cold tolerance have already been isolated from alfalfa and other species (Castonguay et al., 1997; Dhindsa et al., 1998; Thomashow, 1998). Further isolation of genes related to all phases of winterhardiness and dormancy induction are underway in several laboratories. We are attempting to identify genes from various germplasms that are involved in photoperiod sensing, independent of temperature responses. Transformation of diverse germplasm sources of winterhardiness or dormancy-related genes may allow the induction of responses not previously present or the suppression of endogenous genes. Several phenotypes could be manipulation: (i) delaying the photoperiod induction of dormancy responses in subsp. falcata would allow for better fall growth and dry matter yield with little reduction in winterhardiness, (ii) adding photoperiod sensitivity to non-dormant germplasm may greatly enhance its ability to survive the winter, and (iii) altering the various pathways of cold acclimation and winterhardiness in any germplasm could minimize the time needed for plants to acclimate sufficiently for adequate winterhardiness. Our ability to easily manipulate the winterhardiness and dormancy of populations will be a key ingredient in developing useful heterotic groups.
References
Bernardo, R. 1992. Relationship between single-cross performance and molecular marker heterozygosity. Theor. Appl. Genet. 83:628-634.
Bingham, E.T. 1993. Registration of WISFAL alfalfa (Medicago sativa ssp. falcata) tetraploid germplasm derived from diploids. Crop Sci. 33:217-218.
Brouwer, D.J. and T.C. Osborn. 1999. A molecular marker linkage map of tetraploid alfalfa (Medicago sativa L.). Theor. Appl. Genet. [in press].
Brummer, E.C. 1999. Capturing heterosis in forage crop cultivar development. Crop Sci. 39:943-954.
Brummer, E.C., J.H. Bouton, and G. Kochert. 1993. Development of an RFLP map in diploid alfalfa. Theor. Appl. Genet. 86:329-332.
Brummer, E.C., J.H. Bouton, M. Sledge, and G. Kochert. 2000. Molecular mapping in alfalfa and related species. p. In I.K. Vasil and R. Phillips (ed.) DNA-based markers in plants. 2nd. Kluwer, Dordrecht. [in press].
Brummer, E.C., M.M. Shah, and D. Luth. 1999. Re-examining the relationship between fall dormancy and winterhardiness in alfalfa. Crop Sci. [in review].
Busbice, T.H. and C.P. Wilsie. 1965. Fall growth, winter hardiness, recovery after cutting, and wilt resistance in F2 progenies of Vernal x DuPuits alfalfa crosses. Crop Sci. 5:429-432.
Castonguay, Y., P. Nadeau, S. Laberge, and L.-P. Vézina. 1997. Changes in gene expression in six alfalfa cultivars acclimated under winter hardening conditions. Crop Sci. 37:332-342.
Charcosset, A., M. Lefort-Buson, and A. Gallais. 1991. Relationship between heterosis and heterozygosity at marker loci: a theoretical computation. Theor. Appl. Genet. 81:571-575.
Daday, H. 1964. Genetic relationship between cold hardiness and growth at low temperatures in Medicago sativa. Heredity 19:173-179.
Dhindsa, R.S., A.F. Monroy, V. Sangwan, W. Kawczynski, and E. Labbé. 1998. Low temperature signal transduction during cold acclimation of alfalfa. p. 15-28. In P. Li and T.H.H. Chen (ed.) Plant cold hardiness: Molecular biology, biochemistry, and physiology. Plenum, New York.
Diwan, N., J.H. Bouton, G. Kochert, and P.B. Cregan. 1999. Mapping simple sequence repeats (SSR) DNA markers in diploid and tetraploid alfalfa. Theor. Appl. Genet. [in press].
Dudley, J.W. 1964. A genetic evaluation of methods of utilizing heterozygosis and dominance in autotetraploids. Crop Sci. 4:410-413.
Echt, C.S., K.K. Kidwell, S.J. Knapp, T.C. Osborn, and T.J. McCoy. 1994. Linkage mapping in diploid alfalfa Medicago sativa
L. Genome 106:123-137.
Grivet, L., A. D'Hont, D. Roques, P. Feldmann, C. Lanaud, and J.C. Glaszmann. 1996. RFLP mapping in cultivated sugarcane (Saccharum spp.): Genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142:987-1000.
Kehr, W.R., C.C. Lowe, and H.O. Graumann. 1961. Use of first- and advanced-generation two-clone synthetics as parents of four-clone synthetics of alfalfa. Crop Sci. 1:355-358.
McCaslin, M., D. Brown, H. Deery, and D. Miller. 1990. Fall dormancy and winter survival in alfalfa: variation within and between populations. Proc. 32nd North American Alfalfa Impr. Conf. Pasco, WA. 19-24 Aug. NAAIC, Beltsville, MD.
McCaslin, M. and W. Helming. 1995. Examining the use of various factors to predict alfalfa persistence in Wisconsin. Proc. 24th Central Alfalfa Impr. Conf. Spearfish, SD. 18-20 June. South Dakota State Univ.
Ritter, E., C. Gebhardt, and F. Salamini. 1990. Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics 125:645-654.
Rotili, P., T.H. Busbice, and Y. Demarly. 1996. Breeding and variety constitution in alfalfa: present and future. Proc. Grassland and land use systems. . 15-19 September 1996. .
Rowe, D.E. and R.R. Hill, Jr. 1981. Inter-population improvement procedures for alfalfa. Crop Sci. 21:392-397.
Schwab, P.M., D.K. Barnes, and C.C. Sheaffer. 1996. The relationship between field winter injury and fall growth score for 251 alfalfa cultivars. Crop Sci. 36:418-426.
Tavoletti, S., F. Veronesi, and T.C. Osborn. 1996. RFLP linkage map of an alfalfa meiotic mutant based on an F1 population. J. Hered. 87:167-170.
Thomashow, M.F. 1998. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 118:1-7.
Wu, K.K., W. Burnquist, M.E. Sorrells, T.L. Tew, P.H. Moore, and S.D. Tanksley. 1992. The detection and estimation of linkage in polyploids using single-dose restriction fragments. Theor. Appl. Genet. 83:294-300.
Tables and Figure
Table 1. Forage dry matter yields (g plant-1) for the heterosis F1 mapping population and its parents for the first two harvests in 1999 at two Iowa locations, Ames and Nashua.
|
|
Dry Matter Yield (g plant-1) |
|
First Harvest 1999 |
Second Harvest 1999 |
|
Entry |
Ames |
Nashua |
Ames |
Nashua |
|
ABI408 |
70 |
53 |
39 |
13 |
|
Wisfal-6 |
69 |
57 |
25 |
6 |
|
F1 mean |
102 |
99 |
39 |
14 |
|
F1 maximum genotype |
222 |
187 |
66 |
26 |
|
F1 minimum genotype |
10 |
~0 |
6 |
~0 |
|
|
|
|
|
|
Contrast |
Probability level |
|
F1 mean vs ABI408 |
0.0148 |
0.0003 |
NS |
NS |
|
F1 mean vs Wisfal-6 |
0.0312 |
0.0011 |
0.0056 |
0.0001 |
|
ABI408 vs Wisfal-6 |
NS |
NS |
0.0452 |
0.0211 |
Table 2. Broad sense heritabilities (h2)and genetic (rA) and phenotypic (rP) correlations in the F1 mapping population evaluated at two Iowa locations, Ames and Nashua, for fall plant height (October 1998) and winter injury (April 1999).
|
Parameter |
Fall Ht. (± SE) |
Winter Injury (± SE) |
|
 plot basis plot basis
|
0.29 ± 0.035 |
0.39 ± 0.040 |
|
 entry-mean basis entry-mean basis
|
0.69 ± 0.044 |
0.73 ± 0.039 |
|
|
Correlation (± SE) of Fall Height and Winter Injury |
|

|
-0.16 ± 0.048 |
|
 entry-mean basis, Ames entry-mean basis, Ames
|
-0.044 (P = 0.53) |
|
 entry-mean basis, Nashua entry-mean basis, Nashua
|
-0.082 (P = 0.25) |
Fig. 1. Comparison of linkage group 4b from the diploid alfalfa map (Brummer et al., 1993) and linkage groups from each of the parents of the tetraploid mapping population. Asterisks indicate that these markers showed distorted segregation in the diploid F2 population; these markers do not exhibit deviation from the expected 1:1 segregation for SDRF in the tetraploid F1 population. Traits associated with these markers by ANOVA were for first and/or second cut dry matter yields in 1999
Previous Page
 plot basis
plot basis entry-mean basis
entry-mean basis![]()
![]() entry-mean basis, Ames
entry-mean basis, Ames![]() entry-mean basis, Nashua
entry-mean basis, Nashua