D. Z. Skinner
USDA-ARS, Manhattan, KS
The variation in the chloroplast constitution of alfalfa ( Medicago sativa) that has been reported may be classified into one of the five categories of: (1) variation in the number of chloroplast per cell; (2) multiple forms of the chloroplast genome in the same plant (heteroplasmic plants); (3) variation in the intergenic spacer regions that are consistent within the species; (4) insertions and deletions of introns that are consistent within species; (5) and hypervariable regions, probably intergenic, that vary in length within the species and sometimes within the same plant. Molin et al. (1982) measured the effect of ploidy on several traits in alfalfa. Using two series of isogenic lines, Molin et al. determined that the number of chloroplasts per cell was approximately double in a doubled diploid (tetraploid) relative to the original line (mean number of chloroplasts = 21.4 and 40.2, respectively). In a doubled tetraploid (octoploid) the number of chloroplast per cell was again approximately double (31.2 versus 59.1, respectively). However, the amount of chlorophyll per chloroplast was reduced from 5-7 percent in the tetraploid and octoploid, suggesting a compensating effect, tending toward a consistent level of photosynthetic efficiency. The amount of oxygen evolution per cell was approximately double in the doubled tetraploid (4.2 versus 8.1 pmol O2h-1cell-1 ) but somewhat less than double in the doubled tetraploid (8.3 versus 13.9 pmol O2h-1 cell-1). This result suggest the functional capacities of chloroplasts are not identical in different ploidy backgrounds. Presumably, successful 2x by 4x crosses would result in a collection of somewhat different chloroplasts within the same plant, i.e., heteroplasmic plants.
Heteroplasmy in alfalfa was first conclusively demonstrated with molecular techniques by Johnson and Palmer (1989). In that study, it was demonstrated that XbaI digests of some individual alfalfa plants had a single DNA fragment that hybridized to a particular chloroplast DNA probe, while other individual plants had that one fragment plus an additional fragment. Subsequently, Fitter and Rose (1993) demonstrated that the two forms of the XbaI fragment survived with differential efficiency through tissue culture. Fitter, Thomas and Rose (1996) then demonstrated that the two forms of the XbaI fragment differed at a single base within the XbaI site. This XbaI site also was used in a study of meiotic transmission of chloroplast DNAs (Masoud, Johnson and Sorensen, 1990).
Variation of chloroplast genomes also has been identified as insertions and deletions within intergenic regions. Aldrich et al. (1988) studied the region between the psbA and trnH genes in the chloroplast genomes of several plant species. Using Marchantia polymorpha as an ancestral "standard" it was found that alfalfa contains an insertion of 210 bp comprising an inverted repeat stem-loop structure 5' to trnH. Portions of this structure were found in other legumes (Pisum sativum and Glycine max). The differences between alfalfa and these other legumes apparently were due to deletion events, suggesting that alfalfa may be ancestral to some of the other leguminous species. These findings suggest variation in intergenic regions in the chloroplast may be useful in defining evolutionary relationships.
Somewhat similar to the evolution of the psbA/trnH intergenic region, the single intron of the chloroplast rpoC1 gene has been described as variable (present or absent) among genera and among species within genera (Downie et al, 1996). Again using M. polymorpha as an ancestral "standard" it is generally accepted that angiosperm ancestors did have the intron (Downie et al., 1996). Throughout evolution, the intron has been lost independently in several lineages - most notably the lineages leading to the grasses and the Cactoideae. In a survey for the presence/absence of the intron in 107 species representing 54 families, Downie et al. (1996) included alfalfa and seven other Medicago species. Surprisingly, M. suffruticosa subsp. leiocarpa lacked the intron, while all other representatives of Medicago had the intron. Similarly, all other examined representatives of the Fabaceae had the intron. As Downie et al. indicate, this finding distinguishes M. suffruticosa subsp. leiocarpa from all other Medicago, but is uninformative as to the proper evolutionary placement of this species.
The coding regions of chloroplast genomes of higher plants are highly conserved across species and genera. However, some regions of the chloroplast genome have been described as highly variable in several species. In alfalfa, Johnson and Palmer (1989) described individual alfalfa plants that appeared to have multiple forms of the chloroplast DNA molecule (heteroplasmic plants), and also described a genomic region that was hypervariable between Medicago species and showed variation among three alfalfa plants. The variation reported by Johnson and Palmer was found in HaeII digestion products. Schabel (1996) reported a short hypervariable region of the alfalfa chloroplast DNA that showed extreme variation in HindIII digests of alfalfa plants.
Both of these studies (Johnson and Palmer, 1989;Schabel, 1996) used RFLP analysis to observe the variation. Alignment of these regions with the putative gene map developed by Johnson, Ding and Palmer (this conference report) indicated the hypervariable regions occur within intergenic regions. PCR primers have been developed for the regions identified by Johnson and Palmer and by Schabel. Amplification of these areas has yielded evidence of substantial variation (eg. Figure 1). Amplification of multiple bands from the DNA of single plants suggested heteroplasmy originating in these areas is common (Figure 1; lanes 2,3,5,12, 15, 18).

Figure 1. A gel containing the PCR products from a hypervariable region
of the alfalfa chloroplast genome. The region was first described from
HindIII fragments by Schabel, 1996.
To investigate the molecular basis of the hypervariability, 20 ramets of each of two genotypes of 'Riley' were grown in the field for four years. PCR amplification of the hypervariable regions revealed no changes in the HaeII region, but revealed one ramet of one genotype had undergone a deletion in the HindIII region (Figure 2).
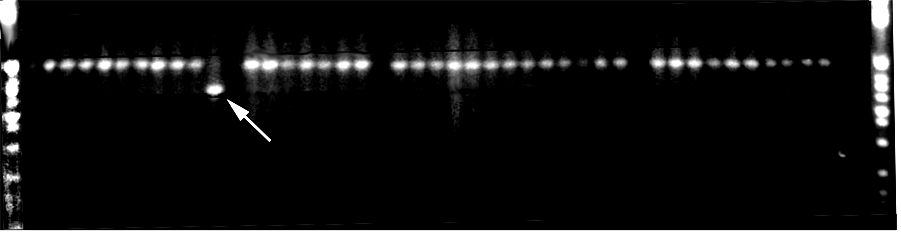
Figure 2. A gel containing the PCR products from 20 ramets each of two 'Riley' genotypes field-grown for four years. The arrow indicates one ramet which has undergone a deletion in a hypervariable region of the chloroplast genome.
The normal fragment and the shortened fragment were cloned and sequenced.
The results indicated the deletion represented a precise excision of 145bp,
with no perturbations to the rest of the sequence (Figure 3).
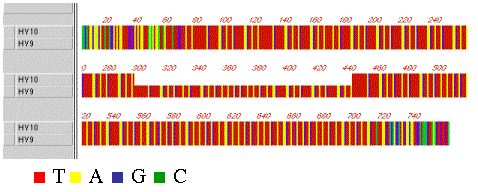
Figure 3. Representation of the DNA sequence of a normal (HV9) and a deletion (HV10) fragment from the hypervariable region of the alfalfa chloroplast genome. The gap in the HV10 sequence indicates the deleted segment.
This finding indicates that the hypervariable regions of the alfalfa chloroplast genome are unstable through vegetative growth; in this one genotype, one deletion was detected in 80 "plant years" of growth (20 ramets grown for 4 years). The molecular basis of variants longer than the original has not yet been determined.
Although the chloroplast is generally thought to be very stable, it now has been demonstrated that portions of the genome are very unstable, and that multiple forms of the genome often exist within the same plant. This kind of variation may account for some of the non-Mendelian variation sometimes reported in alfalfa.
References